|
MotifSuite provides a platform for de novo regulatory motif detection using a stochastic motif detection algorithm with various
motif assessment tools. The suite revolves around MotifSampler, a de novo motif detection tool based on Gibbs sampling that searches for an overrepresented motif in a set of coregulated input sequences. In addition to the core sampler, MotifSuite provides tools to automatically merge the results of multiple stochastic sampling runs and to perform downstream analyses.
NEW (2012-2014): (read more in complementary functionalities in MotifSuite)
- MotifSuite offers two independent and complementary extensions of MotifSampler that perform a motif search in coregulated sequences of a given species extended with orthologous sequences from related species with expected conserved regulatory pathways. The first extension (PHMS) allows identifying conserved or slightly mutated motifs in datasets of related species from one clade. The other extension (NOMS) has the capability to detect mutated or even replaced motifs in distantly related species from a different clade.
- Each of the motif detectors in MotifSuite is equipped with the capability to use additional position-specific prior (PSP) information that can come from any type of regulatory evidence (also epigenetic information) from in vivo, in vitro or computational source. The use of a conservation-based PSP in combination with MotifSampler is most suitable for the detection of motifs with a well conserved motif concensus in other species.
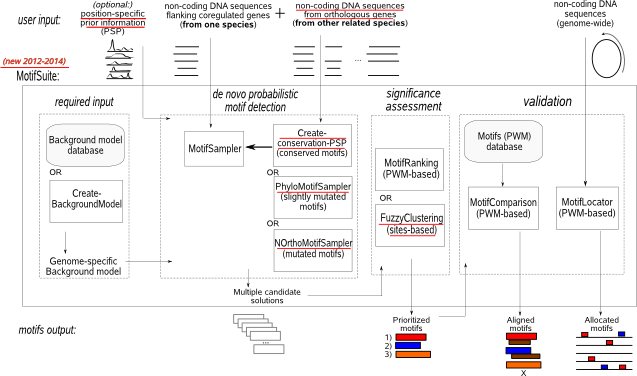
When used as an integrated flow, MotifSuite guides you through the whole process of de novo motif detection going from the selection of a background model, the actual Gibbs sampling, summarizing the output of multiple Gibbs sampling runs, till screening the remainder of the genome with the retrieved motif models (Fig.1, read more in MotifSuite overview). Alternatively, each application within MotifSuite can be used separately and has its own entry page. Submission will initiate the
software command on our server where you can download the applications'output files from.
Optionally, a stand-alone executable of each application can be downloaded for use on your own platform.
The suite consists of the following functionalities:
- generation of a genome-specific background model which can be selected from our database or compiled using CreateBackgroundModel.
- de novo motif detection by means of MotifSampler or its extensions PhyloMotifsampler (PHMS) and NOrthoMotifSampler (NOMS). MotifSampler is a Gibbs sampling based de novo
motif detection algorithm that performs multiple searches for candidate regulatory motifs. Its extensions (PHMS and NOMS) serve to search for motifs that are phylogenetically conserved, slightly mutated or even replaced in related species. All samplers can use position-specific prior (PSP) information from any type of regulatory evidence to guide the computational search to DNA regions that are more likely involved in active regulation. The output of MotifSampler and its extensions is a long list of redundant and not necessarily all relevant solutions.
- prioritizing motifs: we provide tools to postprocess the output of multiple runs of a Gibbs sampling algorithm (or any other stochastic motif detection algorithm). MotifRanking reorganizes the multiple motif detection solutions (PWMs) in a shorter list of non-redundant motifs sorted by their motif score. FuzzyClustering constructs ensemble consensus motifs by analyzing the multiple motif detection solutions at their instance level.
- comparing motifs: MotifComparison analyzes if your detected motif (PWM) corresponds to any of previously described motif models reported in curated databases or detected by yourself in previous analyses.
- genome-wide screening: MotifLocator scans a given set of genome-wide sequences for potential novel instances of your detected motif.
|

 To: Our group ; Contact us ; citing MotifSuite
To: Our group ; Contact us ; citing MotifSuite