|
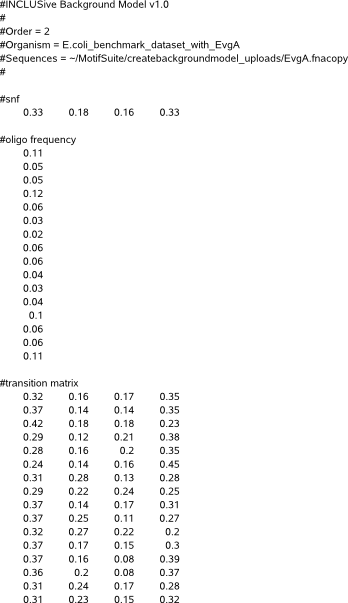
The file starts with a comment line (#INCLUSive Background Model v1.0) that refers to our program and serves as a file recognition for our applications that load a background model file as input.
Next follows a description of the order of the background model (#order) and two informational fields describing respectively a genome identifier (#organism) and the path referring to the sequences data where the model is extracted from (#Sequences).
The single nucleotide frequencies for A,C,G,T are described by 4 tab separated values (between 0 and 1) on the line following #snf. They represent the probability (Pr) to find the respective nucleotide in the sequence dataset where the background is modelled for, independent of the position of this nucleotide in the sequences.
#snf
Pr(A) Pr(C) Pr(G) Pr(T)
The section following #oligo describes the probability of all possible combinations of the nucleotides A,C,G,T of length equal to the background model order (also called an oligonucleotide) in the sequence dataset where the background is modelled for. The total number of oligonucleotides are printed on separate lines and equals 4 powered to the background model order (e.g. 16 for a second order model). The section starts with the oligonucleotide consisting of all A, followed by oligonucleotides where each next position in the oligonucleotide A is repeatedly replaced by respectively C,G,T. Below example is for a second order background model.
#oligo
Pr(AA)
Pr(AC)
Pr(AG)
Pr(AT)
Pr(CA)
Pr(CC)
Pr(CG)
Pr(CT)
Pr(GA)
Pr(GC)
Pr(GG)
Pr(GT)
Pr(TA)
Pr(TC)
Pr(TG)
Pr(TT)
The higher-order background model is described in the section following #transition matrix. Each line in this section describes the tab separated probabilities (Pr) of finding nucleotide A respectively C, G and T given a set of preceding nucleotides of length equal to the background model order. The total number of lines equals 4 powered to the background model order. The preceding oligonucleotide for the first line consists of all A, and in next lines A is repeatedly replaced by respectively C,G,T on each next position in the oligonucleotide. Below example is for a second order background model.
#transition matrix
Pr(A|AA) Pr(C|AA) Pr(G|AA) Pr(T|AA)
Pr(A|AC) Pr(C|AC) Pr(G|AC) Pr(T|AC)
Pr(A|AG) Pr(C|AG) Pr(G|AG) Pr(T|AG)
Pr(A|AT) Pr(C|AT) Pr(G|AT) Pr(T|AT)
Pr(A|CA) Pr(C|CA) Pr(G|CA) Pr(T|CA)
Pr(A|CC) Pr(C|CC) Pr(G|CC) Pr(T|CC)
Pr(A|CG) Pr(C|CG) Pr(G|CG) Pr(T|CG)
Pr(A|CT) Pr(C|CT) Pr(G|CT) Pr(T|CT)
Pr(A|GA) Pr(C|GA) Pr(G|GA) Pr(T|GA)
Pr(A|GC) Pr(C|GC) Pr(G|GC) Pr(T|GC)
Pr(A|GG) Pr(C|GG) Pr(G|GG) Pr(T|GG)
Pr(A|GT) Pr(C|GT) Pr(G|GT) Pr(T|GT)
Pr(A|TA) Pr(C|TA) Pr(G|TA) Pr(T|TA)
Pr(A|TC) Pr(C|TC) Pr(G|TC) Pr(T|TC)
Pr(A|TG) Pr(C|TG) Pr(G|TG) Pr(T|TG)
Pr(A|TT) Pr(C|TT) Pr(G|TT) Pr(T|TT)
|