To: Main Page

To: Main Page

|
Page contents : |
References |
|
Gama-Castro, S. et al. (2008) RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation, Nucleic Acids Research, Vol. 36, Database issue:D120-4. |
Create benchmark datasets |
|
The benchmark datasets consisting of 43 E.coli sequence datasets each containing a known motif derived from RegulonDB are created as follows: RegulonDB is a database on transcription regulation and operon organization in Escherichia coli. We start with the file 'TF Binding Sites' (http://regulondb.ccg.unam.mx/download/Data_Sets.jsp). For each of 43 distinct transcription factors, we select the first target gene of each transcription unit regulated by this transcription factor, and for each target gene, we select the intergenic region 250 nucleotides upstream and 50 nucleotides downstream of the translation start site (as the transcription start site is often unknown). This gives for each transcription factor a file in FASTA format with a list of DNA sequences (separated by '>') for each target gene. |
Table 1 (motif properties) |
|
The benchmark datasets consist of 43 distinct FASTA files each describing a set of DNA sequences. The known motif hidden in each of these files is described by a PWM and by a set of annotated instances for each of 43 distinct transcription factors. 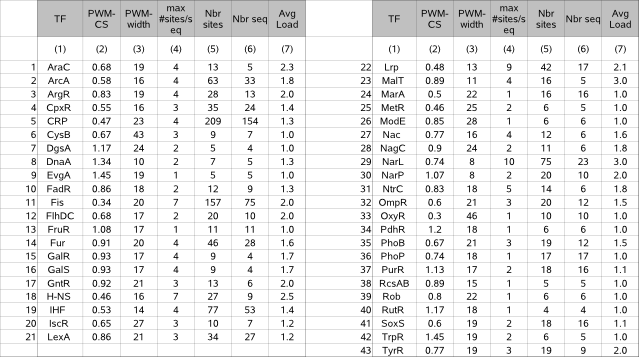 The motif properties in our benchmark lay within the following ranges (exceptions between brackets) : |
Case study: running MotifSuite on the benchmark datasets |
|
The main goal of this case study is to evaluate the motif detection performance of MotifSamplers new release (3.1.5) and to study the pros and contras of the two different prioritizing tools MotifRanking and FuzzyClustering. The results of the case study are discussed in the respective guidelines of each application. This section describes how the applications have been run on the benchmark datasets (input, parameter settings and selection of the output). The workflow presented in the next figure is applied on each dataset and the whole procedure is repeated 10 times to average out particularly good or bad results that may be obtained by chance in the stochastic framework of MotifSampler. 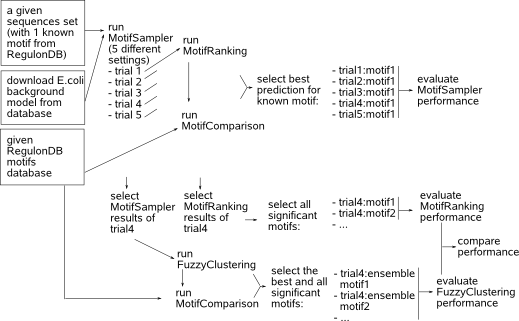 We run MotifSampler in a predefined set of trials that mainly differ in the setting for parameter -p (5 trials as described in table 2a). For each trial, we run MotifRanking with default program parameter settings on the PWM output file reported by MotifSampler to extract different motifs in sorted motif score order and we compare the respective motif models to biologically true motif models in RegulonDB (database available on our server) using MotifComparison. From the list of reported motif models by MotifRanking in each trial, we retain the one with the highest LL score that was detected at least 10 times as the best motif prediction for the known motif in this trial. If no motif with a count higher than 10 was detected in a dataset, we retain the motif model with the yet highest motif count. At this stage, we can evaluate how well MotifSampler finds the known motif and the improvements (e.g. does it find the motif more frequently, is the total number of known instances better described) that can be achieved with the extended design (parameter -p and sampling technique) added in the current release 3.1.5 of MotifSampler. In what follows we further work with the results of MotifSampler obtained in 3.1.5-uniform trial unless MotifSamplers convergence rate was lower than 70, then we choose the results obtained in 3.1.5-fixed trial. |
Computation of performance indicators |
|
The following performance indicators are used throughout this study. Mark that for every performance indicator where the stochastic properties of MotifSampler may influence the result, the reported indicator is averaged over the result in 10 repetitions where MotifSampler was repeated on the same dataset with the same parameter settings. |
Table 2 (MotifSampler performance) |
|
(back to : case study) 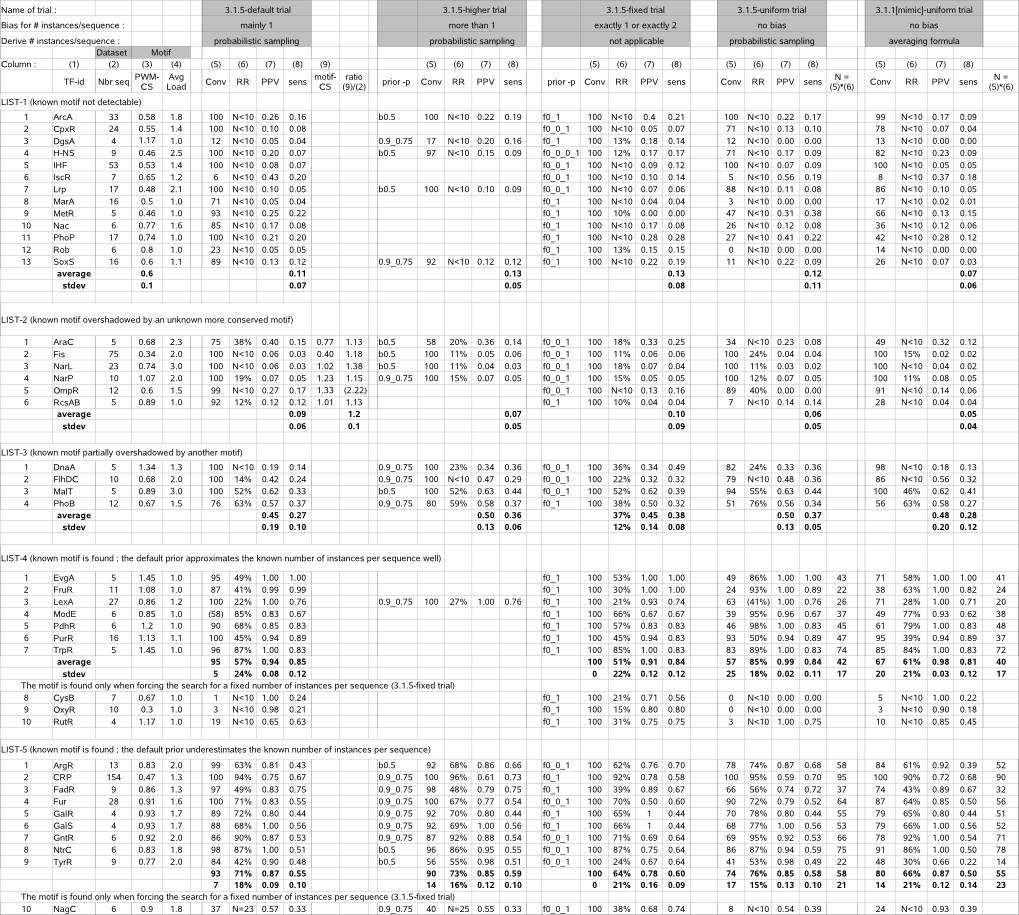
Column (1) : TF = Transcription Factor identifier of a given dataset |
Table 3 (MotifRanking performance) |
|
Table 3 - columns (6-11) list the performance indicators (N, RR, PPV, sens, CS, LL) computed for the candidate true motif(s) (*) retained for a given dataset by running MotifSampler and MotifRanking on 43 benchmark datasets. Columns (2-4) display properties of the dataset and the known true motif, extracted from table 1. The 43 datasets are sorted into 2 lists depending if MotifRanking reported a significant motif (RR and/or N >10) or not. 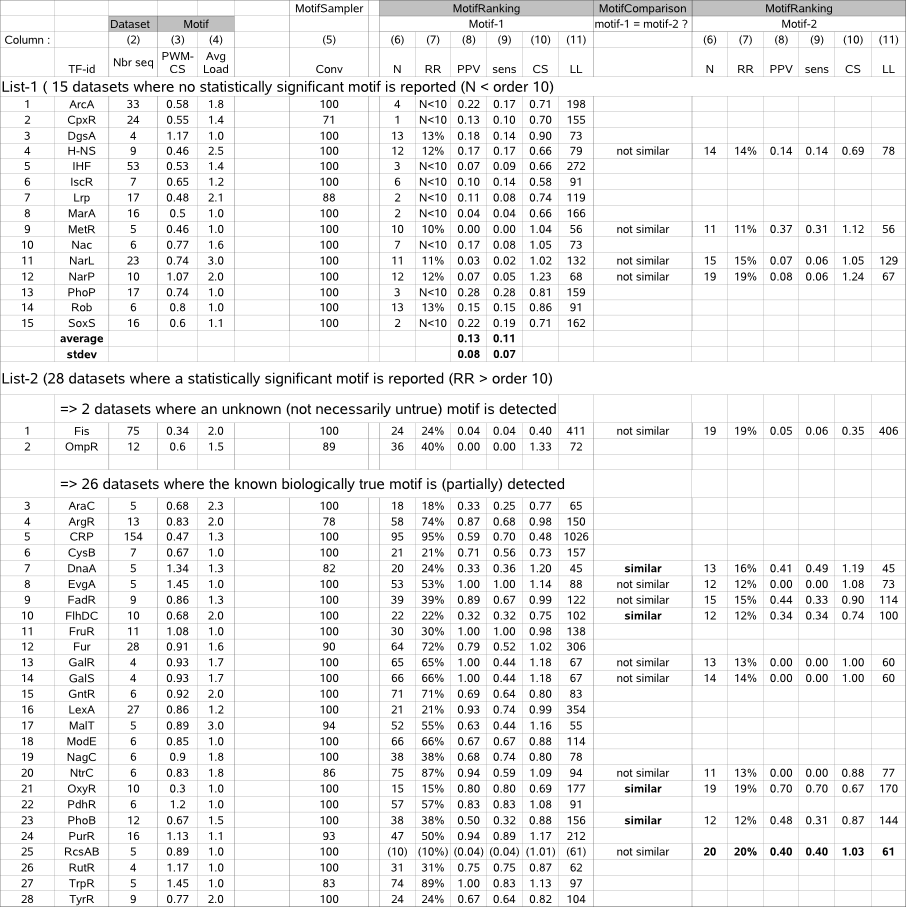
Column (1) : TF = Transcription Factor identifier of a given dataset |
Table 4 (FuzzyClustering performance) |
|
Table 4 - columns (3-8) list the performance indicators (Occ(i), N(m), dM(i), CS, PPV, sens) computed for the candidate true motif(s) (*) retained for a given dataset by running MotifSampler and FuzzyClustering on 43 benchmark datasets. Column (1) displays properties of the known true motif, extracted from table 1. The 43 datasets are sorted into 2 lists depending if FuzzyClustering retained a significant motif or not. 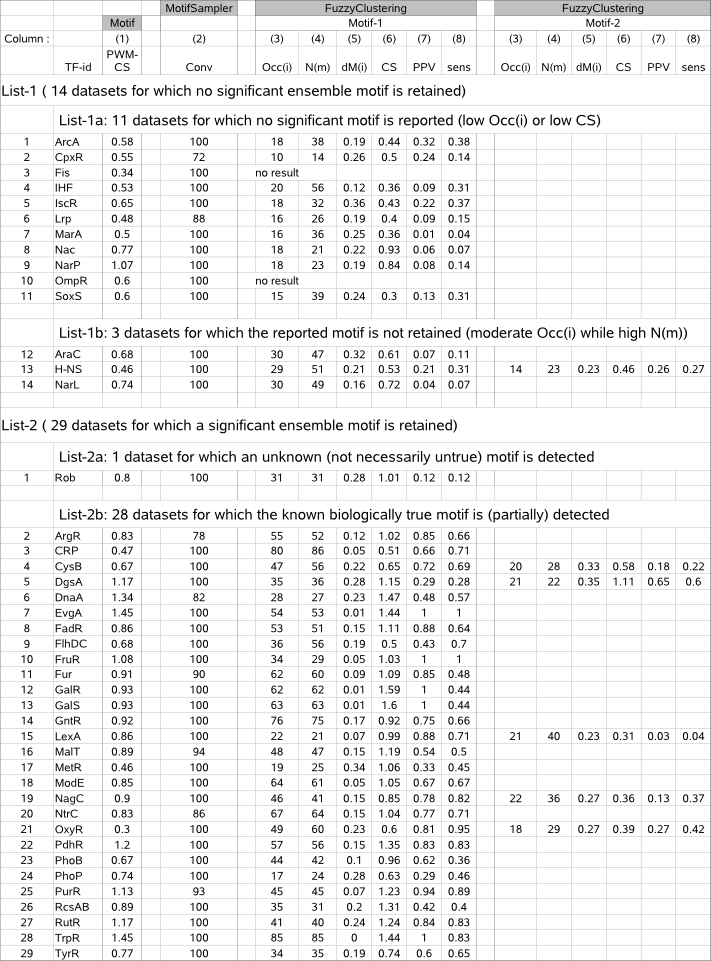
Column (1) : TF = Transcription Factor identifier of a given dataset |