|
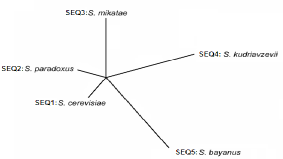
In bioinformatics, NEWICK is a text-based format for representing trees in computer-readable form using (nested) parentheses and kommas. For detailed information how to build such a nested-parentheses tree, we refer to ~/phylip/newicktree.html.
The newick tree occurs on a single line, starting with a greater-than ('>') symbol in the first column and a tree-recognition string, in our case 'Tree' or 'Star', immediateley followed by the (nested) parentheses describing the relations between the species involved in your study:
e.g.
>Star(species1:branchlength1,species2:branchlength2,species3:branchlength3);
or
>Tree(species1:branchlength1,(species2:branchlength2,species3:branchlength3):node_branchlength1);
or with internal node identification (optional):
>Tree(species1:branchlength1,(species2:branchlength2,species3:branchlength3)NODE1:node_branchlength1);
The names of the species involved should be exactly the species-identifiers used in your sequences FASTA file (in this file, the species names also follow the greater than '>' symbol in the sequence identifier lines, see Fasta format). A branchlength consists of integers and (if decimal numbers) a dot (no komma!). Branchlenghts are always preceded by a colon symbol ':'. Internal nodes may but do not need to be identified by a string (we do not further use such node strings in our software). The description of 'NODE1' above is thus optional. The whole line is loaded as one unit, so no white spaces or tabs are allowed in any of the identifiers or numbers or before or after parentheses, kommas or colons.
Mark: The use of a string ('Star' or 'Tree') preceeding the first parenthese is as such required in our software to recognize the newick format, but the choise is not stringent (you can use 'Tree' or 'Star' for both star respectively tree topology, it does not further influence the way of loading the root-nodes-ancestor- and branchlength information).
(expert use, ask for details)
if (pInput[0] == "GeneBias") StoreGeneMutationRates(pInput);
else if (pInput[0] == "CoregBias") StoreCoregulationWeights(pInput);
|