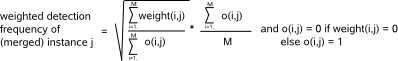The input for FuzzyClustering is a file (parameter -f) containing a list of detected motifs reported by multiple runs of MotifSampler
or any other motif finder in instances format. Each detected motif is described by a motif identifier
(e.g. '#id: motif1' or '>motif1') and a set of instances annotated in a particular sequence set. The annotation of an instance in a sequence describes
the sequence identifier, the start and end position and strand of the instance in the sequence and its nucleotide description e.g. seq13 122 139 +
CTGCCTAC. Optionally, the nucleotide description is followed by a numerical value (instance weight) reflecting the reliability of the detected instance
as assessed by the motif finder (if no weight is supplied by the user, the value is set to 1 by FuzzyClustering).
FuzzyClustering assembles the input of the motif detection results in a matrix F as follows (Fig.1):
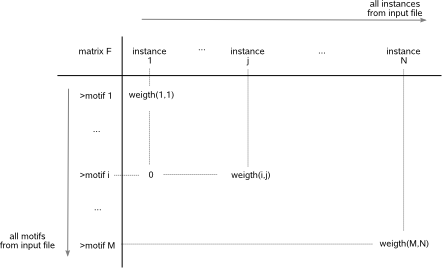
Fig.1: Representation of the input of the motif detection results in a matrix F.
The columns in matrix F represent all instances described in the input file and the rows represent the identifiers of all detected motifs described
in the input file. A matrix entry equals zero when a particular motif does not contain a given instance (e.g. motif i does not contain instance 1), else
it equals weight(i,j): the instance weight of the given instance(j) for this particular motif(i). Subsets of instances that frequently occur together
in subsets of detected motifs will show as sub-matrices (clusters) consisting of mainly non-zero values in matrix F (Fig.2).
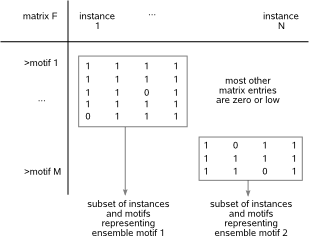
Fig.2: Sub-matrices consisting of mainly non-zero values (clusters) correspond to ensemble motifs.
FuzzyClustering will iteratively extract the most cohesive cluster
that each time corresponds to an ensemble motif by means of a Singular Value Decomposition (SVD, based on
Joshi et al., 2008) of matrix F.
PREPROCESSING OF MATRIX F
A high number of zeros in matrix F complicates the cluster extraction process (SVD) or will decrease the quality of the extracted ensemble motif
(small or in-cohesive clusters). Fig.3 demonstrates how the number of zeros in matrix F drastically increases when detected motifs describe the same motif
on a slightly shifted position compared to using a 'merged' instance that represents all shifted versions of the motif in a given sequence and strand.
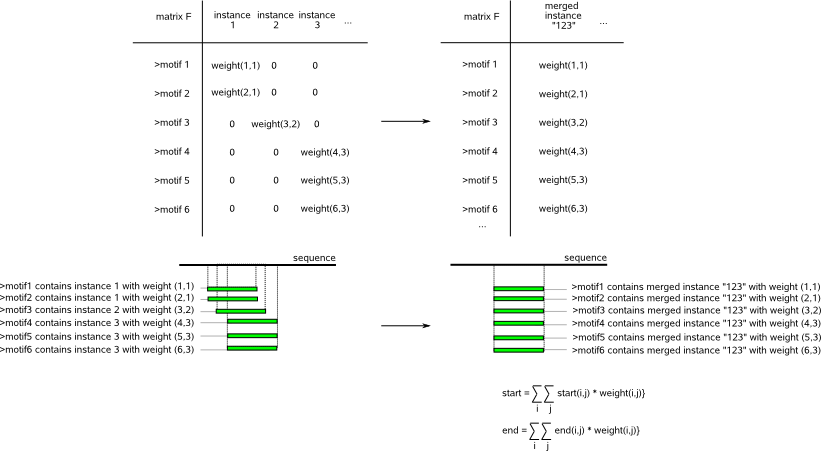
Fig.3: Impact on the sparsity of matrix F when representing shifted versions of the same motif (green rectangles)
in a given sequence/strand as different instances (left) or as one merged instance (right).
On the other hand,
treating overlapping instances as members of the same motif (and thus using a merged column in matrix F) despite them belonging to truly different
motifs that are located in each other's neighborhood on the sequence, will overestimate the clustering signal of a wrongly merged motif. As a trade off, all instances
that overlap for at least 50% with the most frequently detected instance amongst a set of overlapping instances are treated as belonging to the same
motif.
For further analysis, overlapping instances of the same motif are represented by one 'merged' instance (see Fig.3, right) with a start and end
position that equals the average of the start and end positions of the overlapping instances that this merged instance represents
(each instance contributes proportional to its instance weight (weight(i,j)) in the computation of the average start and end).
In matrix F, all columns that correspond to overlapping
instances of a same motif are replaced by one merged column that now corresponds to the merged instance.
Instances (merged) that have a detection frequency (defined in Fig.4) lower than a threshold set by parameter -p, also correspond to columns in matrix F
with many zero values. These spurious instances are removed from the input as they might interfere with the clustering of the more important instances
(those with a higher weighted detection frequency).
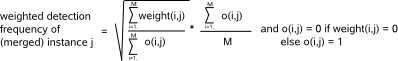
Fig.4: The contribution of each instance in the computation of the detection frequency of a merged instance j is proportional to its instance weight
(= weight(i,j) in matrix F). Formula : the first factor equals the average of the weights of all instances that were merged in instance j, the second
factor equals the number of occurences (sum o(i,j)) of (shifted versions) of instance j divided by the number of detected motifs (M) in the input file.
EXTRACTING MOTIFS FROM MATRIX F (Fig.5)
Applying SVD on matrix F results in the extraction of the most cohesive cluster of instances and detected motifs from matrix F that all correspond to the
same ensemble motif. Each instance and each motif in the extracted cluster get assigned a score (membership). The membership is a relative
measure that reflects how well an instance (motif) represents the ensemble motif compared to the other instances (motifs) in the extracted cluster. The higher the memberships of all instances and motifs in the cluster, the more confidence we can have
in the reliability of the ensemble motif. Instances (motifs) with a low instance (motif) membership are removed from the cluster as they are considered to
represent unreliable predictions that are not sufficiently well supported by the data. An instance (motif) membership is found low when it is lower than
a fraction (set by threshold) of the highest instance (motif) membership observed in the cluster. In default mode, the instance and the motif fraction
threshold are (separately) computed by the program as the value that best balances the size of the cluster (number of remaining instances (motifs) in
the cluster) with the overall reliability of the cluster (coherence of all instance (motif) memberships in the cluster). In non-default mode, the
fraction threshold (in %, applicable for both instance and motif membership) is set by parameter -m.

Fig.5: A cluster extracted from matrix F consists of a set of instances and motifs that all represent the same motif. Each instance and motif is prioritized by
a membership score. Only instances and motifs with a membership score higher than an internally computed (or user-defined) membership threshold will be considered.
Before extracting a second cluster (ensemble motif), each entry in matrix F that corresponds to an instance and a motif that were both assigned to the
first cluster is set to zero in matrix F. Note that instances (motifs) assigned to a previously extracted ensemble motif can still contribute to a subsequent
motif (because only assigned instance-motif entries are set to zero, not the full column (row) of an assigned instance (motif)), referring to the
'fuzzy' aspect of the clustering algorithm. The cluster (ensemble motif) extraction process continues until no more clusters can be extracted
(matrix F becomes too sparse to apply SVD).
POSTPROCESS EXTRACTED MOTIFS
>From the ensemble motifs extracted by FuzzyClustering, only the ones that are expected to represent true motifs are reported in the output.
First, we consider as true motifs those that are overrepresented in most sequences of the input sequence set (assuming the input sequences are choosen
based on high coregulatory evidence): a cluster is only retained if the instances contributing to
the cluster occur in a minimal fraction (as set by parameter -i, default 50%) of the sequence set
(note that the sequence set is defined by all sequence identifiers listed in the input file supplied to FuzzyClustering - this sequence set is not
necessarily the same as the one supplied to MotifSampler as there may be sequences in which no motif was detected by MotifSampler).
Secondly, a cluster must be supported by multiple solutions of the motif finder: a cluster is only retained if the weighted count of contributing motifs (C(m)) to the cluster (computed as the sum of all non-zero motif memberships normalized by the highest motif membership in the cluster) is higher than a minimal
fraction (as set by parameter -j, default 10%) of the number of supplied motifs to FuzzyClustering.
To construct a PWM representation of an ensemble motif, the instances contributing to the cluster are aligned, starting with the most reliable instance
(highest instance membership) and gradually adding instances with lower instance membership with a maximal nucleotide match, minimal shift and
at least 50% overlap to the most reliable instance. The PWM is derived from this alignment as follows: each instance contributes a nucleotide count
for A, C, G or T (or N if the instance is not described on a particular position in the alignment) that is proportional with the instance membership
in the cluster (together with a small pseudocount). We define the core PWM as the one having the same width as the most reliable instance.
This core PWM is further extended to the left and right as far as the alignment reaches and the PWM with the highest CS score is chosen as
the representative of the extracted ensemble motif. Finally, only ensemble motifs for which the representative PWM has a CS score higher than
a minimal setpoint (parameter -c, default 0.5) are reported in the output.
|