PWM stands for Position Weight Matrix and describes the probability to find the respective nucleotides A,C,G,T on each position of a motif.
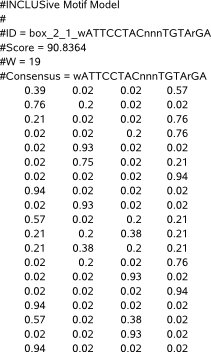
The file starts with a comment line (#INCLUSive Motif Model) that refers to our program and serves as a file recognition for our applications that load a PWM file as input.
Next follows the PWM description of a first motif, starting with some comment lines.
The first comment line describes a unique motif identifier (#ID). The second comment line shows a motif score (#Score) which can be a score that is computed from the PWM or any other score that reflects the importance of the motif being described. The following two lines give the PWM length (#W) and a consensus description (#Consensus) of the motif. A consensus description is derived from the information available in the PWM ; it is a string-based sequence representation of the motif in IUPAC code symbols (A,C,G,T,n,s,w) that describes the most likely nucleotide(s) on each position in the motif (n = any of A,C,G,T, s = C or G, w = A or T).
The comment lines are immediately followed by the values that make up the PWM (matrix) : each line describes the tab-separated probabilities (Pr) for nucleotide A, C, G and T on a given position in the motif. The number of lines must equal the length of the motif (#W).
The probabilities described in a PWM can be frequencies (normalized values between 0 and 1 and the sum of a row equals 1), or they can be represented as counts (values can be higher than 1 and zeros are also common). Our applications in MotifSuite have been designed to work with (and report) PWMs that describe frequencies.
! MARK : decimal numbers in a PWM must be described using a DOT (not a komma) e.g. 0.54 (not 0,54).
Pr(A,1) Pr(C,1) Pr(G,1) Pr(T,1)
Pr(A,2) Pr(C,2) Pr(G,3) Pr(T,4)
...
Pr(A,W) Pr(C,W) Pr(G,W) Pr(T,W)
The motif description ends with a blank line return. The second and following motifs are described in exactly the same way, each time separated from each other by a blank line. The end of the file is recognized by the last blank line return. Note that there is no explicit numbering of the motifs in the file.
Some comment lines may display typical information when reported by a specific application in MotifSuite (this section assumes you have read and understood the guidelines of the respective applications in MotifSuite) :
- #ID in MotifSampler : the motif identifier typically starts with 'box', followed by underscore-separated information consisting of the number of MotifSampler run, the number of detected motif in this run and a consensus representation of the motif.
Example : #ID = box_3_1_ATTCCTACnnnTGTArGA.
- #ID in FuzzyClustering :
the motif identifier consists of underscore('_')-separated fields starting with '#id: box', the number of the reported motif in the file (simple sequential numbering), the symbol 'CS' or 'W' indicating if the reported PWM is the PWM representation with the highest consensus score (CS) respectively the longest motif length (W) and in [brackets] the length, consensus description and consensus score of the motif being described. The end may describe additional information such as (%InstanceCut) the fractional threshold that was used to remove unreliable instance predictions of this motif, (%dataCut) the fractional threshold that was used to remove data-motifs that do not represent this motif, (nbrSeq) the number of sequences with at least one instance of the motif, (nbrInst) the number of instances of the motif and (nbrData) the number of data-motifs that also detected this motif (Mark: data-motifs are the motifs described in the input file supplied to FuzzyClustering, each data-motif represents a motif detected by one run of a motif finder).
Example : #id: box_1_CS[18,ATTCCTACnnnTGTArGA,1.44974]_%InstanceCut=0.34_%DataCut=0.41_nbrSeq=9_nbrInst=9_nbrData=21.
- #Score in MotifSampler : the reported value is the log-likelihood score of the motif being described. The log-likelihood score is not a score that is computed from the PWM. It is a score that is maximized during the motif detection process to find the motif in the dataset that optimally balances motif conservation with the number of motif instances.
- #Score in MotifRanking : the reported score is the score that was used for sorting the candidate motifs in descending motif score order. The type of score that was used is specified with parameter -m in MotifRanking.
- #Score in FuzzyClustering : the reported score is the consensus score (CS) of the PWM, which is a measure for the conservation of the motif.
|