To: Main Page ; To: Application ; To: Guidelines

To: Main Page ; To: Application ; To: Guidelines

Setup of the test dataset |
Setup of the test dataset |
|
The use of a PSP and different types of conservation-based PSPs has been extensively studied in combination with PRIORITY (Gordan et al., 2009). PRIORITY is conceptually similar to MotifSampler in that both are Gibbs sampling overrepresentation based motif finders. In the next case study, we demonstrate that the integration of a PSP in MotifSampler results in a same performance increase as demonstrated for PRIORITY in 62 yeast datasets. At the same time, we demonstrate that the use of a conservation-based PSP is a powerful method to integrate phylogenetic motif conservation into our Gibbs sampling algorithm. |
Performance of MotifSampler with a conservation-based PSP |
|
We run MotifSampler (parameter settings in appendix-1) in each of the 62 datasets in 3 different trials. The first trial does not use any PSP, the second trial uses PSP-C describing positional alignment-free conservation-based prior scores, and the last trial operates with the discriminative conservation-based PSP-DC. 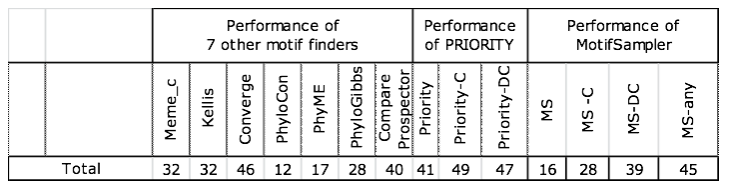
The results show a low number of successes for a non-PSP based detection with MotifSampler (MS). We add two marks that this low performance does not bring down the value of MotifSampler as such. First, only the highest scoring significant motif was retained for further validation on success. We cannot exclude that the target motif for a given dataset was not reported as a lower scoring motif, neither did we verify if the retained motif matched another true motif (other than the target literature motif). Secondly, two other motif finders (PhyloCon and PhyME) obtain a performance of only the same order although they use an extra level of information (conservation in orthologs) in their motif finding algorithms. The low number of successes of MotifSampler is therefore attributed to the abundant presence of local optimal solutions that confound the detection of the target motif. |
Feedback |
|
Contact us if you have comments, questions or suggestions or simply want to react on the contents of this guideline. Thank you. |
References |
- Gordan, R., Narlikar, L., & Hartemink, A. (2009). Finding regulatory DNA motifs using alignment -free evolutionary conservation information. Nucleic Acids Research, 38(6):e90. |