
To: Main Page

This page summarizes the MotifSuite pipelines developed in a recent PhD (Claeys, 2014). |
Justification: |
|
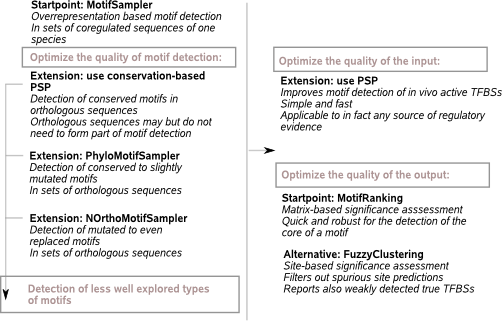
MotifSampler was developed in a previous PhD (Thijs, 2003) and belongs to the class of Gibbs sampling overrepresentation based tools intended for use in the coregulation space. The tool was included in benchmark studies (Tompa et al., 2005, Hu et al., 2005, Singh et al., 2008), forms part of ensemble platforms (Tmod (Sun et al., 2010), MotifLab (Klepper and Drablos, 2013)) and can be valued as a competitive player in the field of computational motif detection. MotifSampler is however prone to reporting local optima (overrepresented solutions that are not involved in regulation): from in-house usage, motifs are typically retained at a significance of order 10 to 20% (assessed by MotifRanking), which does not represent a very convincing strongly overrepresented motif signal. |
Search for more weakly overrepresented motifs: |
|
The basic MotifSampler (Thijs, 2003) is equipped with a probabilistic estimation of number of instances to be annotated per sequence during Gibbs sampling. It works with a prior parameter that biases the motif search to mainly one instance in each sequence in the dataset. Although a higher number is possible, this ‘mainly one’ setting proved to be (and still is) the best to find the core of a motif. Other occurrences are typically retrieved with a motif scanning tool using the detected core motif (MotifLocator). |
Refine significance assessment at the motif instance level: |
|
MotifSampler reports a list of motif predictions obtained from multiple repeats of the motif search initiated at random seeds. A significance assessment of this output is typically done at the motif matrix level using MotifRanking: if the PWM of a high scoring motif is similar to a minimum number of other motif matrices in the list, the motif is found a significant prediction. Other than its moatrix representation, a predicted solution is also reported by the list of sites from which the PWM is constructed. As the input sequences for motif detection in general also contain some noisy sequences (the motif is not present) and different sequences not necessarily contain an equal number of instances, the reported sites list likely still contains some spurious instances that do not belong to the motif. This is especially true if MotifSampler is run with the fixed prior for the number of instances per sequence. |
Search for motifs phylogenetically conserved in related species: |
|
MotifSampler is designed for motif detection in the coregulation space of one species. If run on coregulated sequences from different species separately, each output will be confounded by local optima that do not represent motifs of the common TF for the related species.
Searching in the combined coregulation-orthology space is one remediation to filter out solutions that are not overrepresented in each of the involved species. |
Integrate any type of position-specific regulatory prior information (PSP): |
|
While the present-day computational motif detection algorihtms can accurately predict in vitro binding, solutions represent the in vivo reality more accurately when used in concert with additional knowledge. Integrating additional knowledge into the Gibbs sampling scheme can be challenging with computational or analytical complexities and uncontrolled outcome. The use of a PSP on the contrary, to guide the detection towards prioritized DNA region without excluding this region, is fast, easy and efficient. |
References: |
|
Claeys, M. (2014). PhD: Probabilistic algorithm for finding motifs in sets of orthologous sequences. Leuven: KUL Department Molecular and Microbial Systems. |