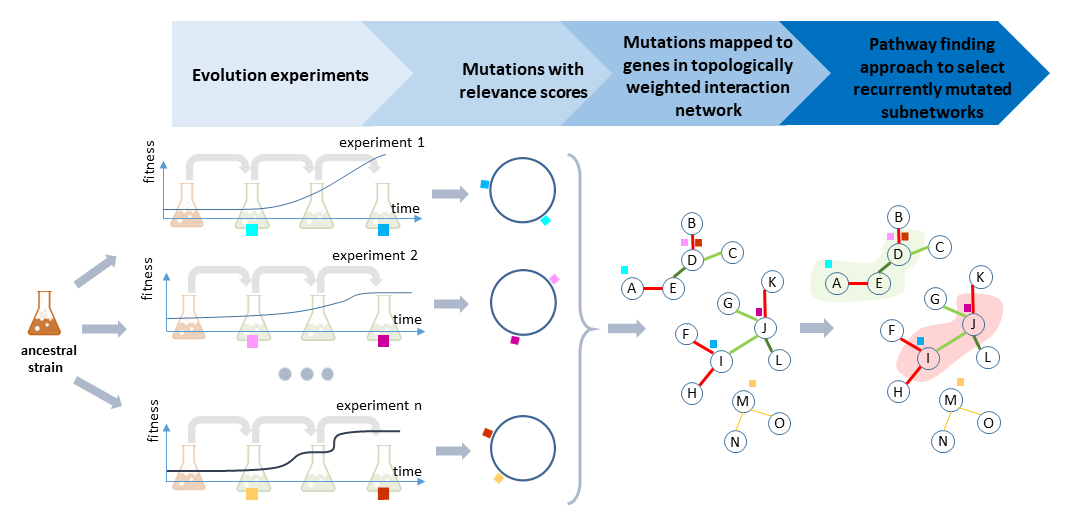
IAMBEE's Analysis Flow
Calculating relevance scores
IAMBEE takes as input the genomics variants of each parallel evolved population before and after an indicated selection sweep. Variants should be called using the ancestral strain as a reference.
IAMBEE will then calculate for each variant a relevance score. This relevance score takes into account:
- The extent to which the variants increases in frequency during the sweep.
- The functional impact of the variant.
- The mutational load of the population in which the mutation occurred.
For an explanation on how these relevance scores are calculated we refer to the extended help file. Users can decide to calculate themselves the relevance scores. For the format in which the relevance scores should be uploaded click here.
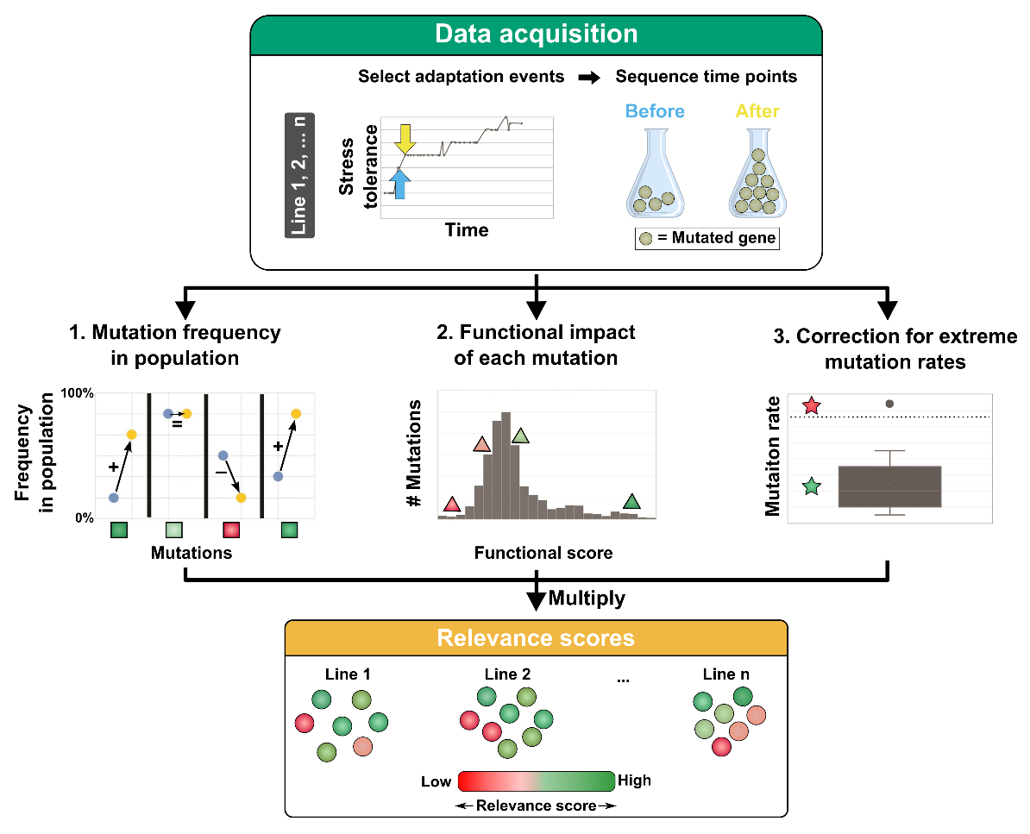
In the data acquisition step, a selective sweep of interest is chosen from an evolution experiment involving multiple parallel evolved populations. Samples taken at time points just before (blue arrows) and just after (orange arrows) this selective sweep are sequenced and mutations are called. For every mutation, a functional impact score and frequency change in the population are determined by the IAMBEE software. The frequency change is derived from the degree to which the mutation changes in frequency before (blue) and after (orange) the selective sweep. Genes with mutations that rise in frequency have higher frequency increase scores (green square) while a low frequency increase score is assig ed to genes with mutations that decrease in frequency in the population (red squares). Next, a functional impact score is assigned to each mutation). Genes with mutations having a high functional impact score are depicted with green triangles and vice versa. In addition, populations with a mutation rate that is significantly higher than the mutation rates of the other populations are detected. The relevance score of mutations that originate from populations with a deviating mutation frequency will be downweighed.
Identifying recurrently mutated sub-networks
To identify recurrently mutated subnetworks, IAMBEE uses a topology weighted interaction network. This network can be uploaded by the user or is available for a selected set of model organisms.
IAMBEE maps all mutations to genes on the interaction network. Each of the mutations (and hence mutated genes) have an assigned relevance score (see above). Given the relevance scores of the genes (nodes in the network) and the weights of the edges (defined by the topology), IAMBEE uses a pathfinding strategy that aims at connecting through the network as many as possible mutated genes with high relevance scores, that preferentially originate from distinct populations. Connections are constrained to be sparse (using the least number of edges). This latter constraint enforces the solution towards a subnetwork of tightly connected genes carrying relevant mutations. This subnetwork is referred to as the adaptive pathway.