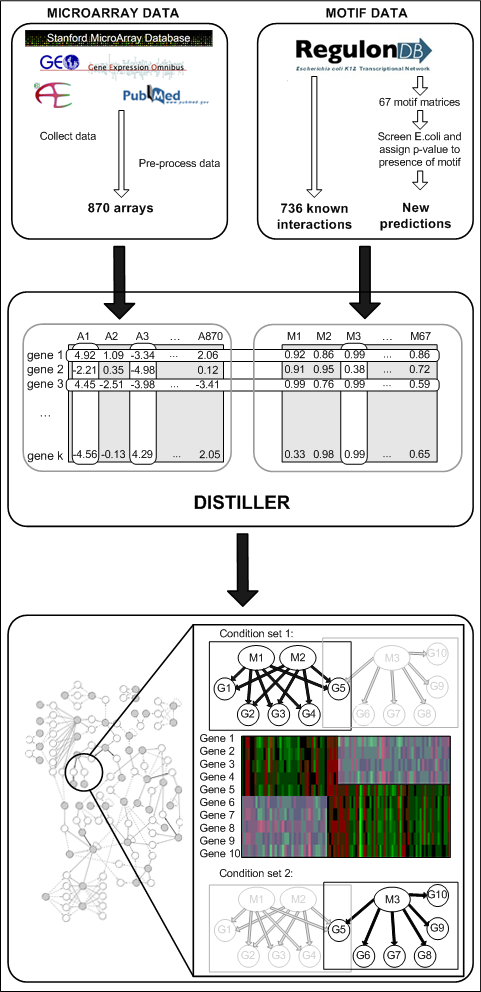
[View full size image]
The framework (software available upon request) builds upon advanced itemset mining approaches that have been designed to have good scalability, efficient memory use, and a small number of user parameters. It includes a condition selection or bicluster strategy in which co-expression of genes is required in only a significant subset of the complete condition set. By including this condition selection we can apply the algorithm to large expression compendia where interesting genes are not necessarily co-expressed in all measured conditions. Our approach also makes it straightforward to include any number of data sources related to transcriptional interactions such as additional microarrays, ChIP-chip or motif data.
Our methodology consists of three steps: (1) the identification of seed modules; (2) the reduction of the set of all seed modules to a manageable set of non-redundant and significant seed modules; and (3) the extension of the thus obtained seed modules with additional genes.
In this study, we applied DISTILLER to simultaneously analyze two distinct data sources: a cross platform expression compendium consisting of 870 microarrays and a regulatory motif compendium consisting of both predicted and experimentally verified motif instances E.coli microarray experiments were collected from the three major microarray databases (Parkinson et al, 2007; Barrett et al, 2007; Demeter et al, 2007) and combined in a cross platform compendium of 870 arrays. From RegulonDB (Salgado et al, 2006), we collected the binding site models (weight matrices) of 67 regulators. These were used to screen E.coli upstream sequences in order to predict potential regulator-target interactions (Hertzberg et al, 2005). Supplementing these predictions with the known gene - motif interactions from RegulonDB resulted in the total interaction matrix.
In summary, DISTILLER identifies modules that consist of genes that are co-expressed in a subset of conditions, together with the controlling regulators. Modules can overlap in regulator, gene and condition content. This allows for identifying condition dependent combinatorial regulation: in this example gene 5 is regulated by both M1 and M2, but under a different set of conditions.