To: Main Page ; To: Guidelines

To: Main Page ; To: Guidelines

|
MotifSampler's program parameter -p specifies Pr(x), the prior distribution on the number of instances of a motif in a sequence. The argument x equals the number of instances of a motif in a sequence and varies from zero to a maximal number set by the user (program parameter -M). Pr(x) is used in each iteration step of the motif detection process to calculate the number of instances of a motif that should be sampled in each sequence (read more in MotifSampler Guidelines, see fig.5). |
Description of 5 types for Pr(x) |
|
We propose 5 models for a prior distribution Pr(x) : |
Mathematical description |
|
Here follows the mathematical description for each type of prior distribution : |
Graphical illustration |
|
Ter illustration follows a graphical representation of some prior distributions at different parameter settings : 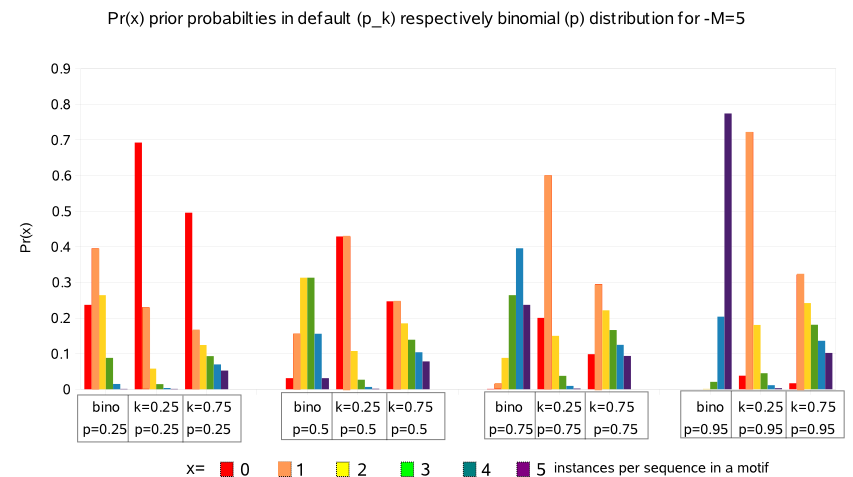 |
Feedback |
|
Contact us if you have comments, questions or suggestions or simply want to react on the contents of this guideline. Thank you. |