SomInaClust
Detection of Cancer Genes Based on Somatic Mutation Patterns of Inactivation and Clustering
About SomInaClust
SomInaClust is a method that identifies cancer driver genes from whole exome/genome somatic mutation data. It specifically prioritizes genes with a higher than expected number of clustering and/or protein-truncating somatic mutations and further classifies them as putative oncogenes (OGs) or tumour suppressor genes (TSGs).
How to install SomInaClust
SomInaClust is implemented as an R package. It can be easily installed from any R console using the following instructions:install.packages("http://bioinformatics.intec.ugent.be/sominaclust/R/SomInaClust_1.0.0.tar.gz", type = "source", repos = NULL)
library(SomInaClust)
If SomInaClust fails to install check the following:
- Do you have the latest R version (SomInaClust needs v3.1.2 or higher)? To check:
version$version
- Do you have the required packages installed? To check and install:
if(!"plotrix"%in%installed.packages()) install.packages("plotrix")
if(!"foreach"%in%installed.packages()) install.packages("foreach")
if(!"doParallel"%in%installed.packages()) install.packages("doParallel")
if(!"parallel"%in%installed.packages()) install.packages("parallel")
How SomInaClust works
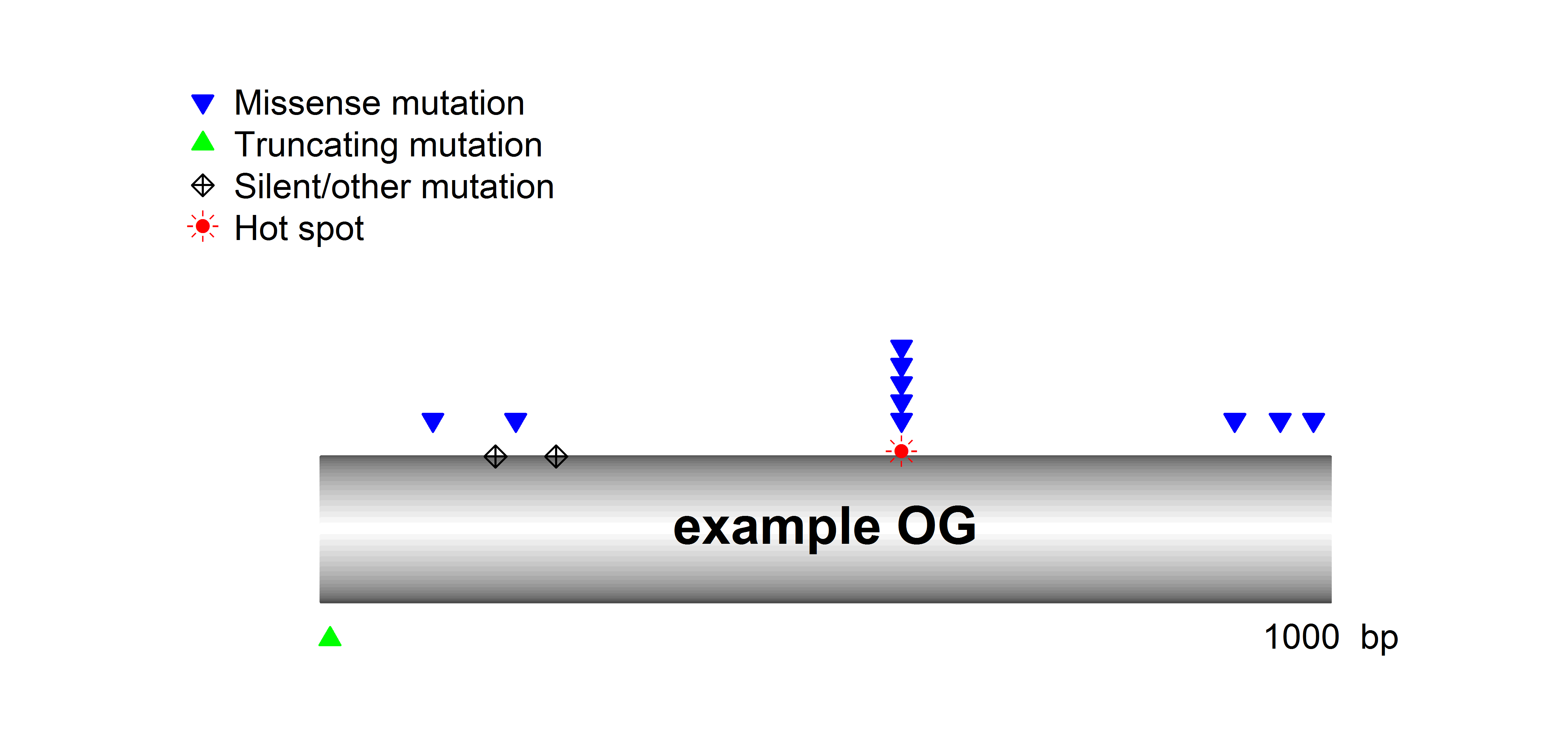
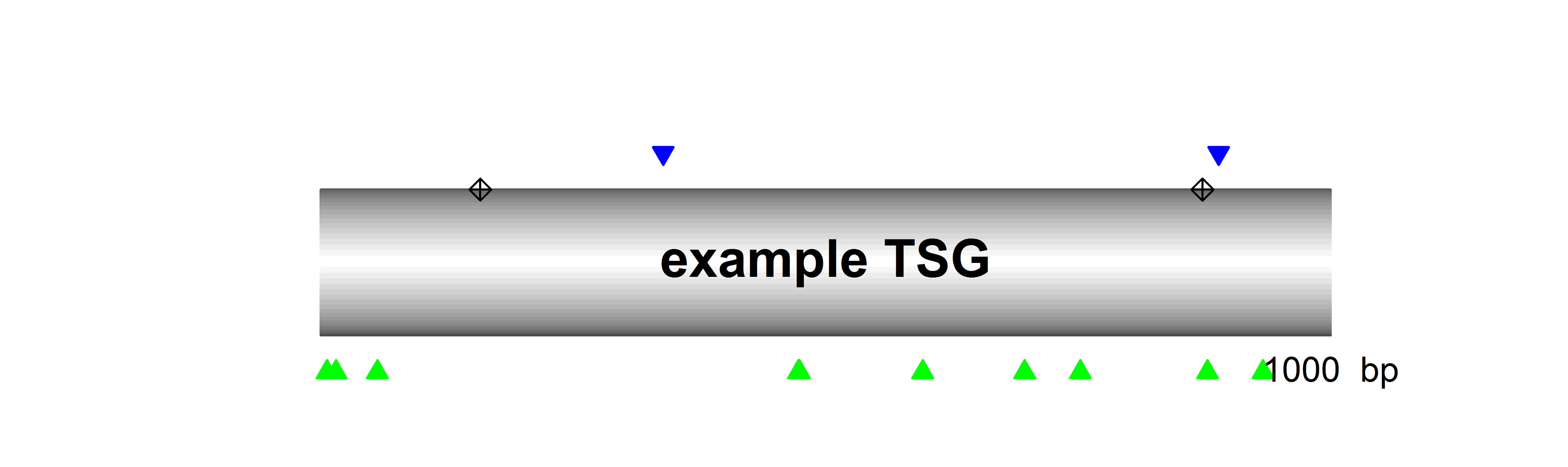
SomInaClust identifies candidate driver genes from whole exome or genome mutation data based on their mutation patterns. The basic assumption is that, due to selective pressure, driver genes are characterized by:
- clustering mutations at hot spots across tumour samples
- a high number of inactivating (protein-truncating) mutations across tumour samples
While the former is the main pattern expected for OGs, the latter is the main pattern for TSGs.
Driver gene identification
Based on both characteristics a gene-specific binomial statistic (qDG) is calculated that allows gene prioritization and the identification of candidate driver genes.
OG and TSG predictions
Based on the proportion of hot-spot located non-truncating mutations on the one hand and the proportion of truncating mutations on the other hand candidate driver genes are further classified as putative OGs of putative TSGs.
More details
For further details on the methodology we refer to the original publication
How to use SomInaClust
The main methods available in the SomInaClust package are
- SomInaClust_det() is used to identify cancer genes from mutation data using precalculated reference files. To view details:
help(SomInaClust_det)
- SomInaClust_ref() is used to calculate the reference files from a reference database. To view details:
help(SomInaClust_ref)
Available data in the SomInaClust package
The following data are provided as part of the SomInaClust package:
- maf_brca
- The maf file that was used to identify the candidate breast cancer driver genes in the original publication is provided as an example file. To view details:
help(maf_brca)
- SomInaClust_results_brca
- Example of the output obtained after running SomInaClust_det on the maf_brca file. To view details:
help(SomInaClust_results_brca)
- ref_v71_clusters
- A dataset specifying for each gene the genome position of the mutation clusters, as calculated using SomInaClust_det on the Cosmic v71 database. To view details:
help(ref_v71_clusters)
- ref_v71_CDSlength
- A dataset specifying for each gene the median CDS length of all transcript variants that are present in the Cosmic v71 database, as determined using SomInaClust_det on the Cosmic v71 database. To view details:
help(ref_v71_CDSlength)
- ref_v71_corr
- A dataset specifying for each gene the OG and TSG correction factors, as calculated using SomInaClust on the Cosmic v71 database. To view details:
help(ref_v71_corr)
- CGC
- CGC list (v71), as downloaded from cosmic and after conversion of gene names to current HGNC symbols. To view details:
help(CGC)
- converttable_mutation_cosmic_tcga
- Table to be used to convert mutation types as defined in cosmic to/from TCGA. To view details:
help(converttable_mutation_cosmic_tcga)
- converttable_genenames_HGNC
- Table to be used to convert gene names to currently approved HGNC symbols. To view details:
help(converttable_genenames_HGNC)
How to cite SomInaClust
Disclaimer
THE SOFTWARE IS PROVIDED “AS IS”, WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.