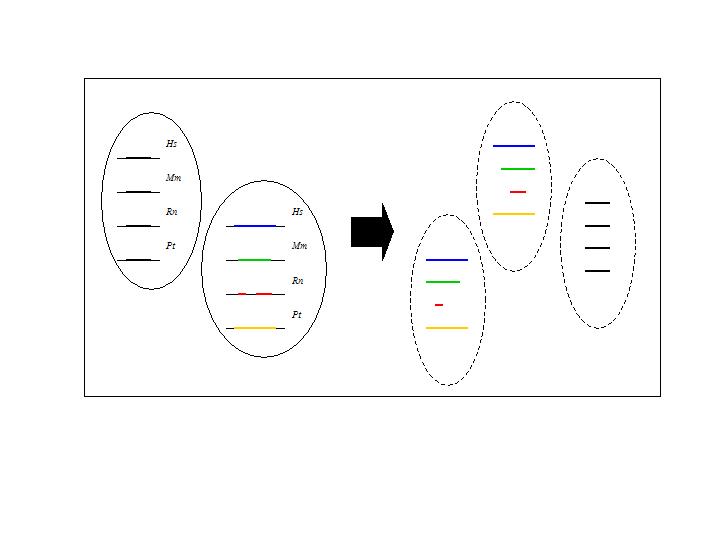
Schematic representation of subclusters, i.e. clusters of conserved orthologous sequences that contain one region in each ortholog.
It is possible that a cluster includes different subsequences originating from the same intergenic region, for instance when multiple smaller subsequences of a certain ortholog show homology with a single longer subsequence of another ortholog. This is the case for the cluster shown in figure 1: it contains a single homologous subsequence of each mammalian ortholog except for the rat for which the cluster contains two subsequences. Both rat subsequences show homology with a single region in the other three mammalian intergenic sequences, but are in the rat sequence separated by a non-conserved gap. Such gaps often result from sequences containing an NNN-stretch that does not match with the other intergenic sequences. Because these separated subsequences map to a different region of the cluster, they are not likely to contain the same motifs. Including them in the data set for motif detection would increase the noise in the motif detection input set. To minimize the noise in data sets used for motif detection, such clusters are split into subclusters. Each subcluster contains exactly one subsequence per ortholog. The number of subclusters that have to be analyzed equals the number of possible combinations of region IDs, for which each ortholog is only included once. Each subcluster, together with the intergenic sequence of the corresponding Fugu ortholog, is then used as input for the subsequent motif detection step.