Benchmark data
The application of chromatin immunoprecipitation combined with DNA microarray techniques (ChIP-Chip) in eukaryotes allows the genome wide mapping of the physical interaction between a TF and its target gene. The test set we composed was derived from a genomewide ChIP-Chip analysis performed by Boyer et al. 2005 [1]. It consists of 116 genes that co-bind three core TFs, OCT4, SOX2, NANOG (involved in plurypotency and self-renewal) in their 1000 bp proximal promoter region. Moreover, the three TFs bind in each other close proximity turning them in a true case example of a CRM.
The 116 out of 353 genes from UCSC [2] that were used in our analysis can be found here. The motif models for the three transcription factors OCT4, SOX2 and NANOG that were downloaded from TRANSFAC [3] can be found here, while the motif models for the 584 PWMs from Transfac [3] can be found here. The EnsemblIDs [4] for the 5000 randomly human genes can be found here
Comparison of ModuleDigger with other CRM detection tools
We compared ModuleSearcher, Clover and ModuleDigger on our benchmark data set. We used all 116 intergenic sequences as input. The number of input PWMs was varied. We started with the easy case in which we only provided four PWMs as input (the three PWMs known to belong to the CRM and one randomly sampled from TRANSFAC). Then we gradually increased the complexity of the problem i.e, finding the right CRM by using more TFs as input.
The 10 sets of PWMs that were used in the analysis with four and ten TFs can be found here
The results of the CRM detection methods on our benchmark data set can be found
here
Results of ModuleDigger on the benchmark data set
For benchmarking ModuleDigger we fed in the 1000 bp proximal promoter regions of all 116 genes. We considered only those modules that contain the combination of all three TFs, i.e. OCT4, SOX2 and NANOG, as true modules. All other modules were not considered to be true.
The performance of the algorithm is assessed by the average rank a true module receives after running the algorithm. In our test we started off with the simple case in which we only used 10 TFs as input (the three true ones together with one randomly sampled one). The complexity of the problem was increased by gradually including more randomly selected TFs (20, 30 or 40 TFs).
The 10 sets of PWMs that were used in the analysis with 10 TFs, 20 TFs, 30 TFs or 40 TFs can be found here
Result of ModuleDigger with 10 TFs, 20 TF, 30 TFs and 40 TFs on our benchmark data can be found:
Result Table
Analysis of the significance of the module ranking by using order statistics
We used the method of Tusher et al. [5] and compared the score of the true module received with the score of a module that received a similar rank as our true module, in the randomized dataset. A true module with a score higher than an equally ranked module in the randomized dataset was considered significant. We also assessed the number of false positive modules that are expected to be present in the module: false positives were defined as the number of modules in the randomized dataset that contained a score higher than the score of the true module in our benchmark dataset.
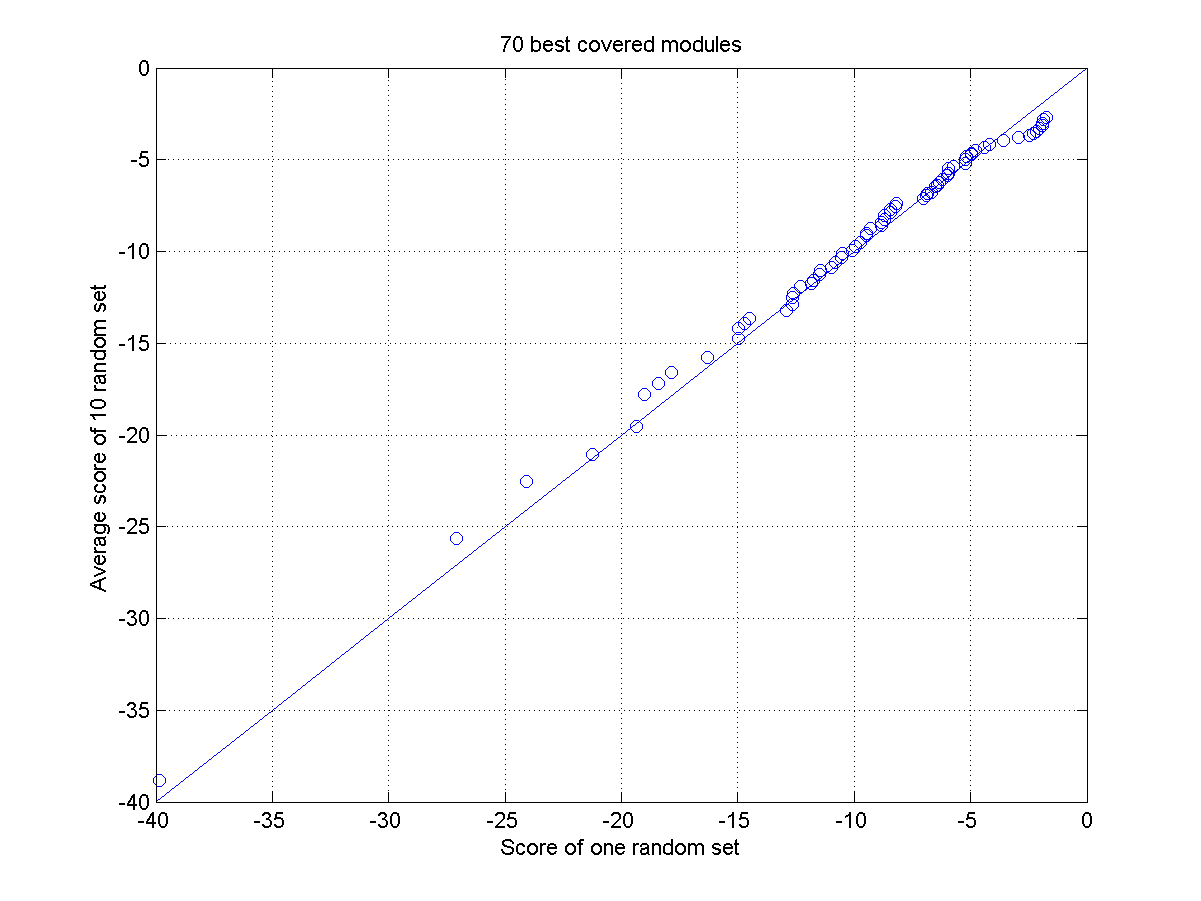
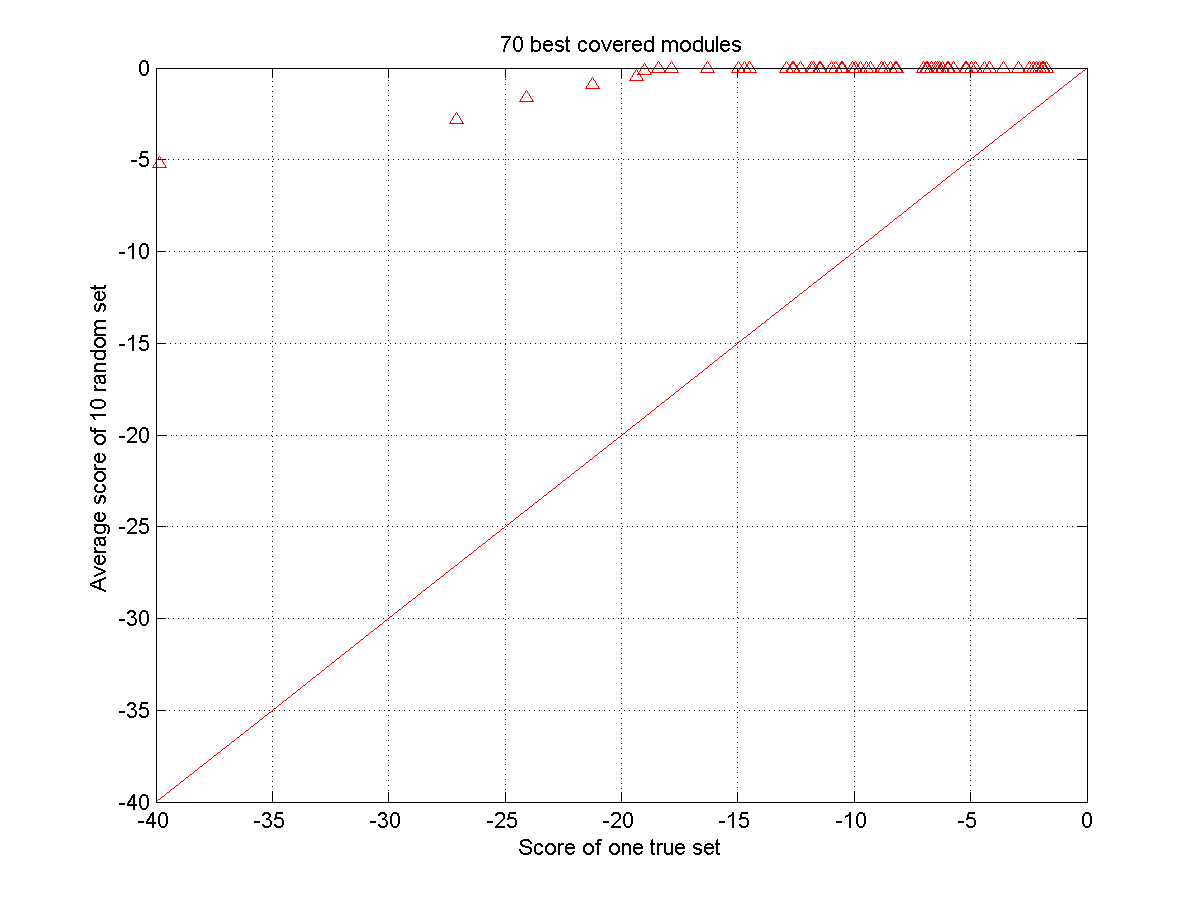
Plot of the scores of 70 best ranked modules versus a baseline for a) a random set and b) the true set. The baseline consists of the average scores of the 70 best ranked modules in 10 different randomizations. For the true sets we used as input a set of 10 TFs amongst which OCT4, SOX2 and NANOG were present, the random sets use as an input a set of TFs without the OCT4, SOX2 and NANOG. The random sets are thus not expected to contain any true modules. Panel a) all selected modules are random and reflect scores of false positives. They are distributed on a one-one line. Panel b) the scores of the highly ranked modules in the true dataset score consistently higher than the equally ranked modules in the random sets.
 |
 |
| [View full size image] | [View full size image] |
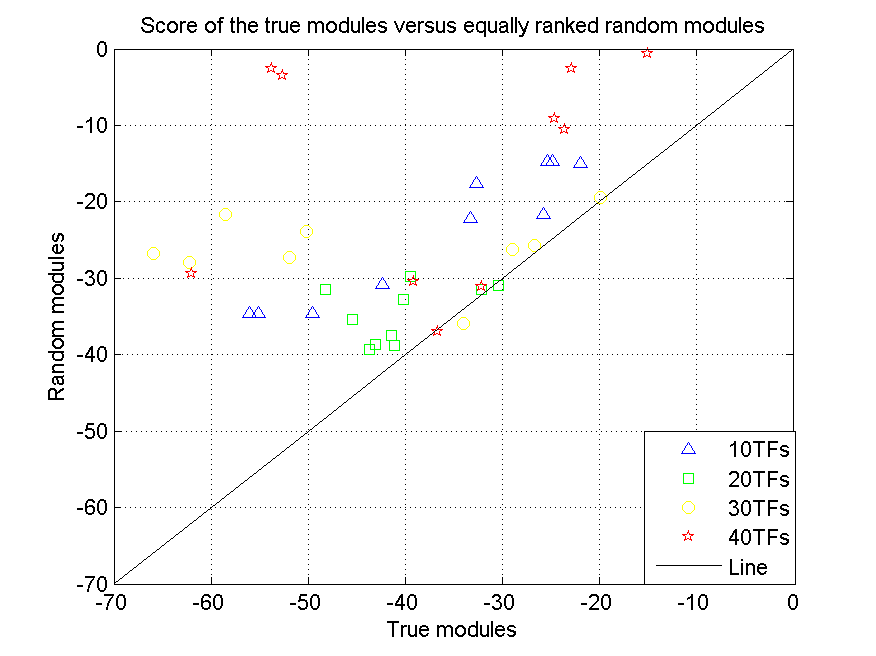
For each true module its score (log value, the lowest one is the best) is plotted versus the score (log value) of the equally ranked module in the randomized dataset. Different symbols correspond to the different datasets of increasing complexity (using respectively 10, 20, 30 and 40 TFs as input).
 |
| [View full size image] |
Download and Manual
ModuleDigger runs on Linux and is free to use for academic purposes only.
For any non-academic purpose, please contact the corresponding author.
ModuleDigger and manual can be download here.
Citation
If you use ModuleDigger in your research, please cite the following publication:
Sun H, De Bie T, Storms V, Fu Q, Dhollander T, Lemmens K, Verstuyf A, De Moor B, Marchal K. ModuleDigger: an itemset mining framework for the detection of cis-regulatory modules (2009). BMC Bioinformatics, 10(Suppl 1):S30, doi:10.1186/1471-2105-10-S1-S30.
