Number of conditions
Number of seed genes
Number of extended seed genes
Correlation extended seed
No. (putative) regulators in module
=
=
=
=
=
75
5
9
0.8859
1
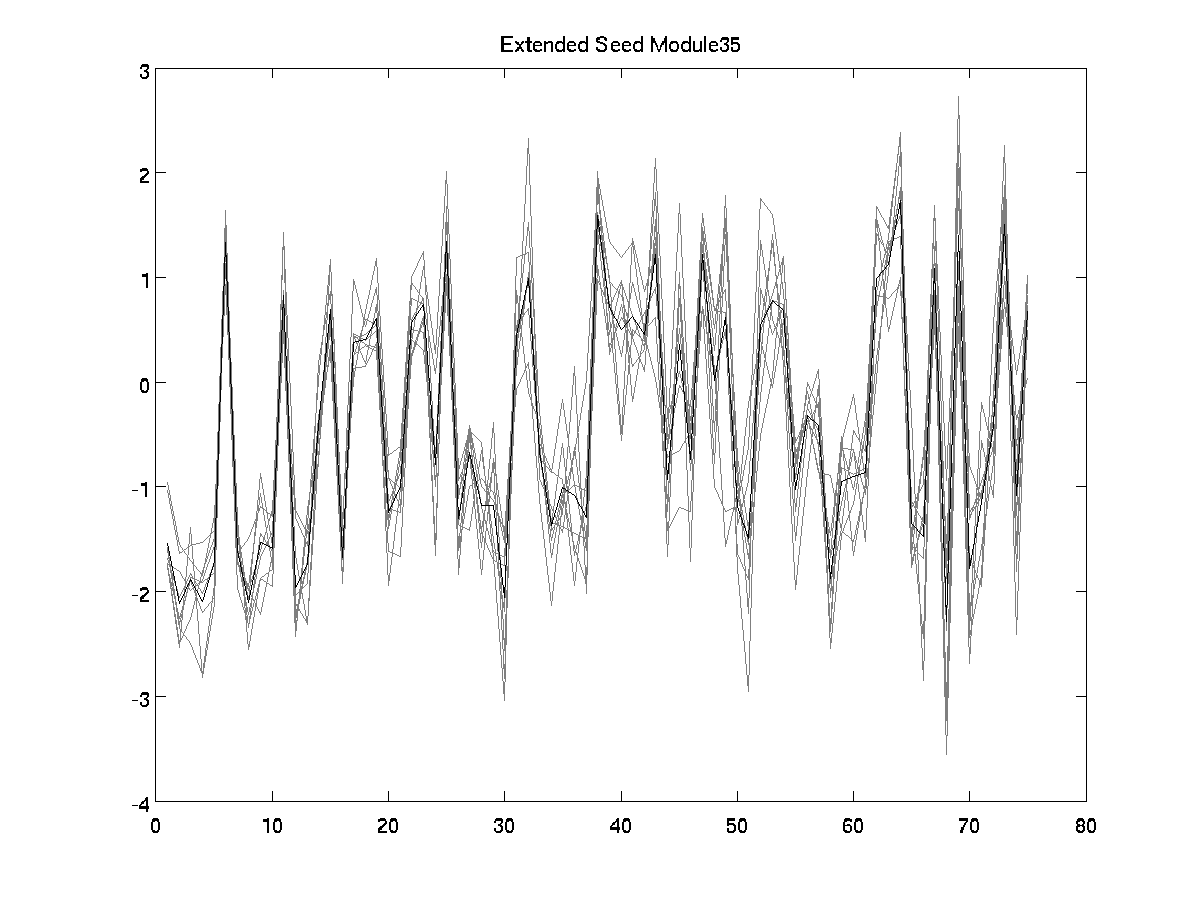
[View full size image]

[View full size image]
| Number of motifs Number of conditions Number of seed genes Number of extended seed genes Correlation extended seed No. (putative) regulators in module | = = = = = = | 1 75 5 9 0.8859 1 |  [View full size image] |  [View full size image] |
| Motif | Motif logo | Module motif logo |
|---|---|---|
| Fis: Activates ribosomal RNA transcription. Plays a direct role in upstream activation of rRNA promoters | 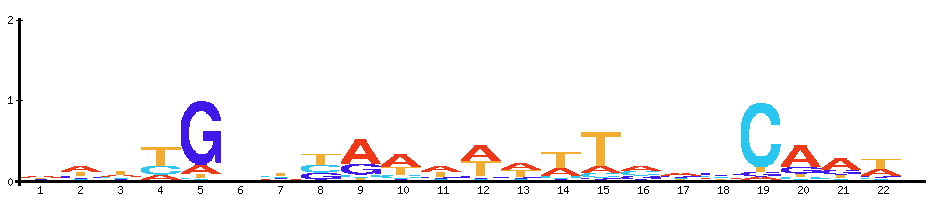 |  |
| Gene | Seed/Extended Seed | Description | Direct interaction (RegulonDB) | Indirect interaction (RegulonDB) | Motif instance |
|---|---|---|---|---|---|
| b3260 (dusB) | Seed | tRNA dihydrouridine synthase | | - | ATTTGCCGATTATTTACGCAAA |
| b3965 (trmA) | Seed | tRNA(m5U54)methyltransferase | Fis | ? | ATCTGGACATCTGATGAGCAAT |
| b1274 (topA) | Seed | DNA topoisomerase I | Fis | + | ATTTGCTTGACATTCGACCAAA |
| b0835 (yliG) | Seed | predicted SAM-dependent methylase | | GTATGGCTATTTTTTGAGCAAA | |
| b0405 (queA) | Seed | b0405 | Fis | + | CATCGATTATATTCTATCCAAA |
| b3261 (fis) | Extension | Fis transcriptional dual regulator | | - | |
| b1344 (ttcA) | Extension | predicted C32 tRNA thiolase | | GTTTGCCCCCAATTTGCCCCAT | |
| b0469 (apt) | Extension | Apt | | AGGCGGTCAAAGGTTAAACAAC | |
| b1876 (argS) | Extension | | CGTTGCTGACGAAAACAGCCAT |