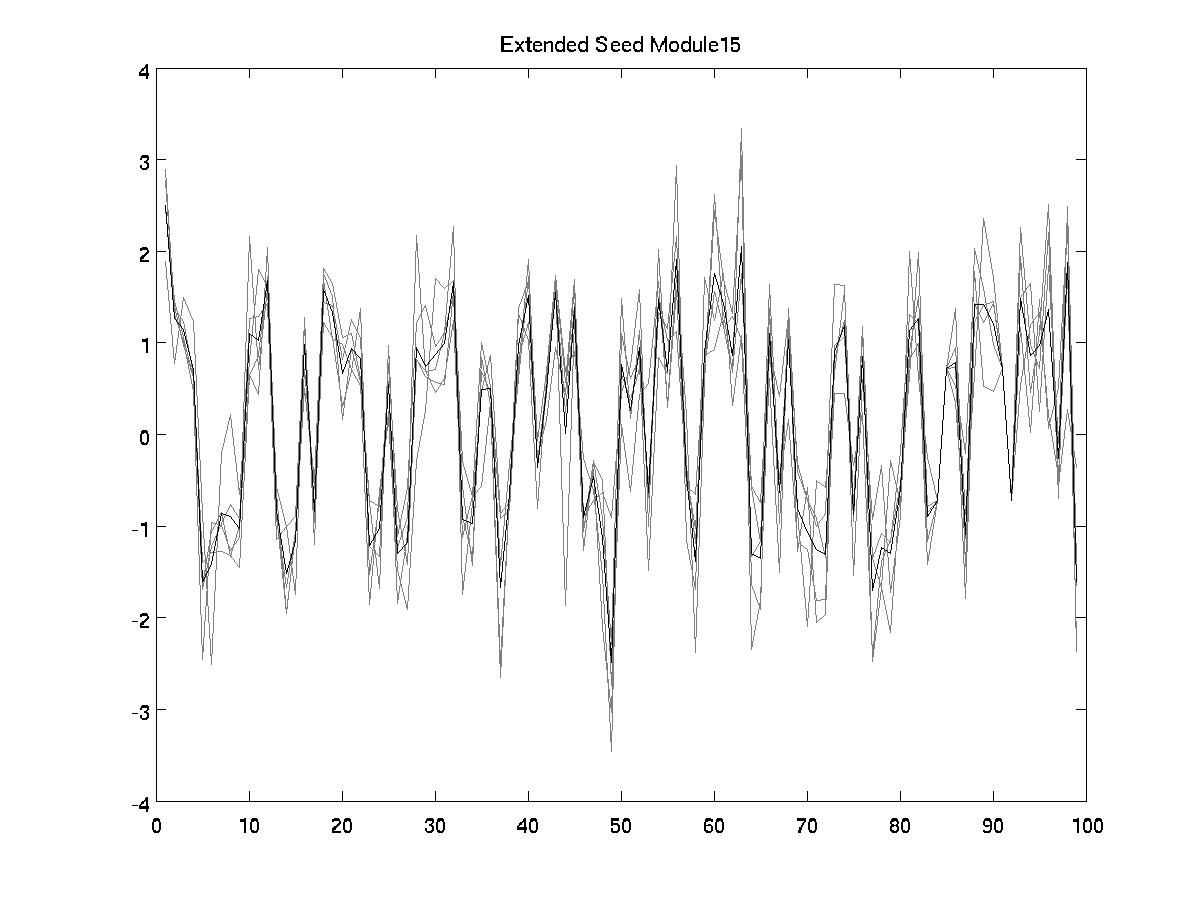
Number of conditions
Number of seed genes
Number of extended seed genes
Correlation extended seed
No. (putative) regulators in module
=
=
=
=
=
99
4
5
0.90728
2
[View full size image]

[View full size image]
| Number of motifs Number of conditions Number of seed genes Number of extended seed genes Correlation extended seed No. (putative) regulators in module | = = = = = = | 2 99 4 5 0.90728 2 |  [View full size image] |  [View full size image] |
| Motif | Motif logo | Module motif logo |
|---|---|---|
| Fis: Activates ribosomal RNA transcription. Plays a direct role in upstream activation of rRNA promoters | 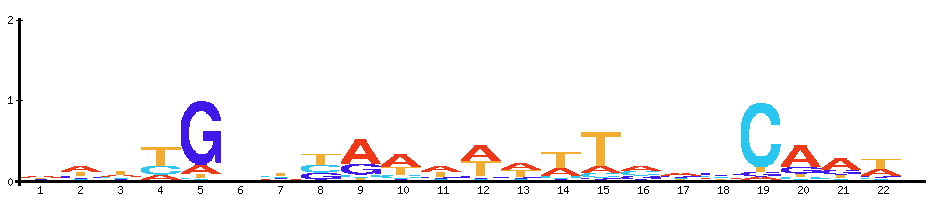 |  |
| CRP: cyclic AMP receptor protein | 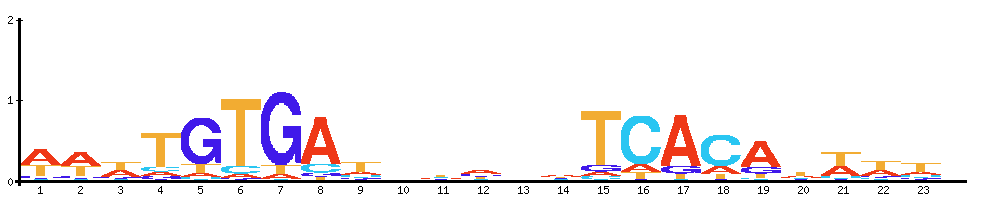 |  |
| Gene | Seed/Extended Seed | Description | Direct interaction (RegulonDB) | Indirect interaction (RegulonDB) | Motif instance |
|---|---|---|---|---|---|
| b4000 (hupA) | Seed | Transcriptional dual regulator HU-α (HU-2) | Fis CRP | + + | GTAAGGATAACTTATGAACAAG AAACGTGATTTAACGCCTGATTT |
| b0440 (hupB) | Seed | Transcriptional dual regulator HU-β, NS1 (HU-1) | Fis CRP | - + | TAAATCTCAATTGATCGACAAG AAGTGCGATATAAATTATAAAGA |
| b1241 (adhE) | Seed | AdhE | Fis | + | TGATTTTCATAGGTTAAGCAAA AAATTTGATTTGGATCACGTAAT |
| b4142 (groS) | Seed | GroES, chaperone binds to Hsp60 in pres. Mg-ATP, suppressing its ATPase activity | | TTGCGCCCAAATTTTGGGCAAC AAATGTGAGGTGAATCAGGGTTT | |
| b1779 (gapA) | Extension | glyceraldehyde 3-phosphate dehydrogenase-A monomer | CRP | + | AGTCGCGTAATGCTTAGGCACA AATCGTGATGAAAATCACATTTT |