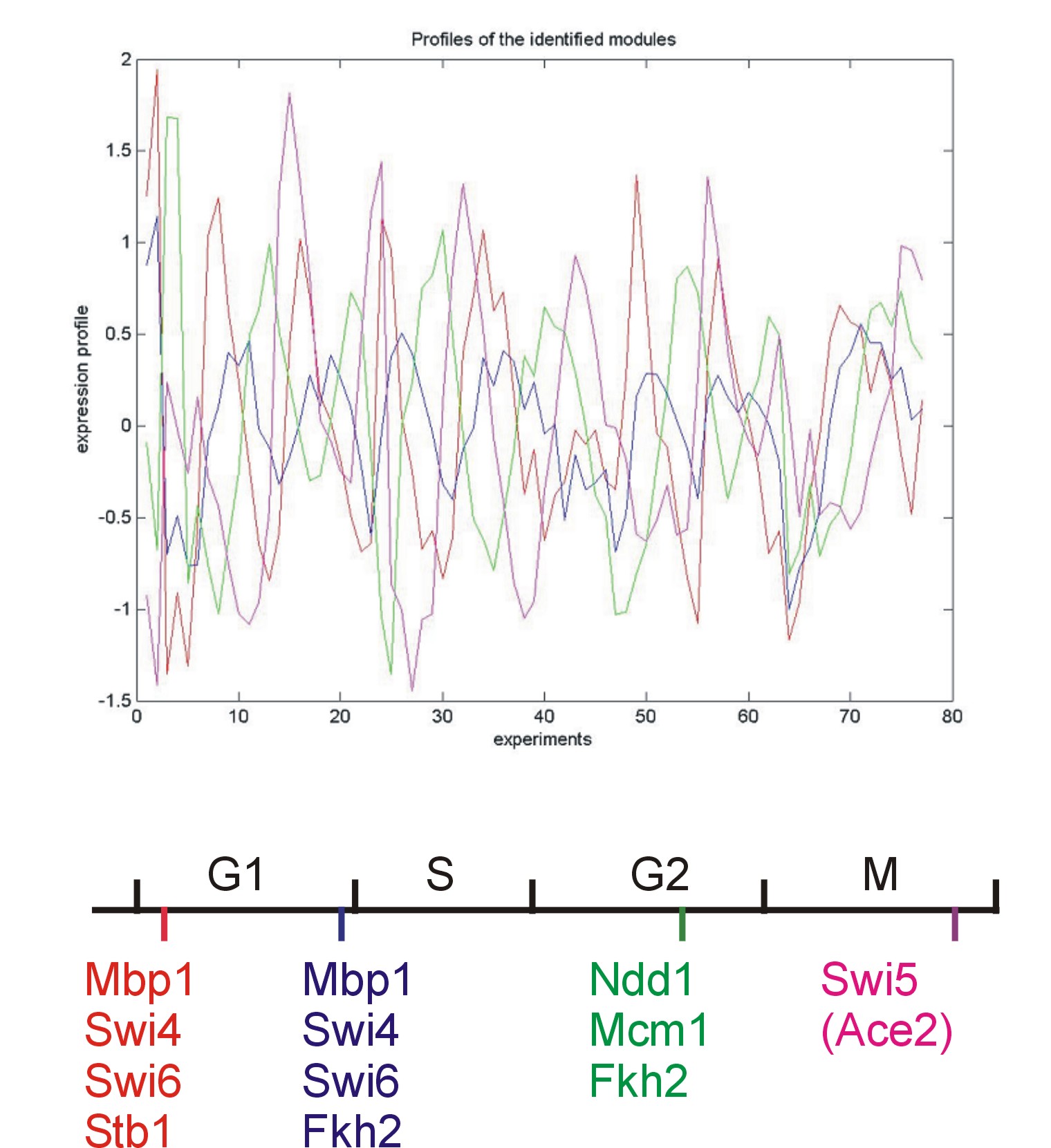
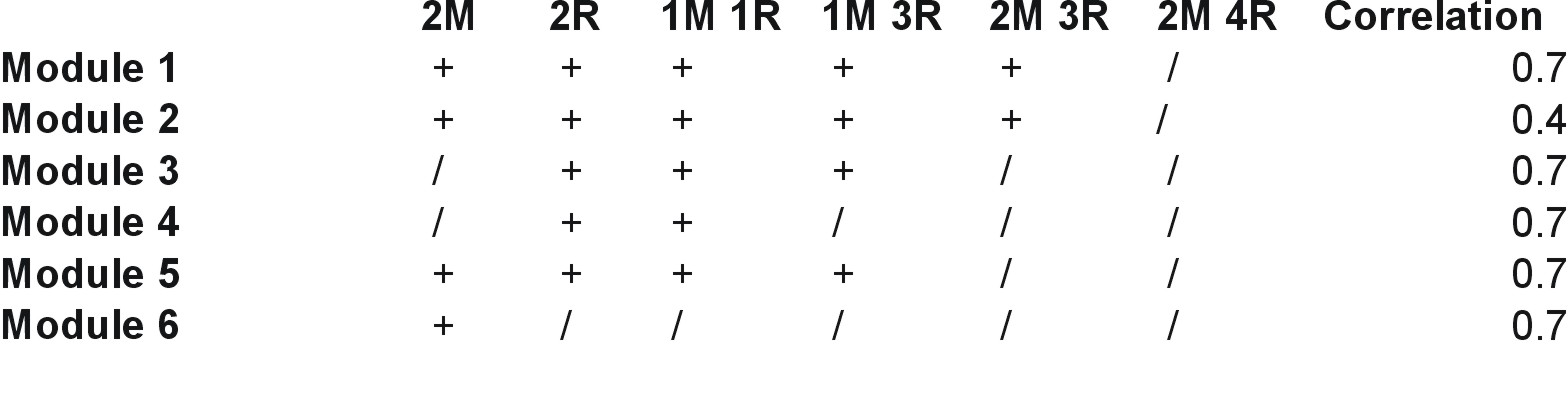
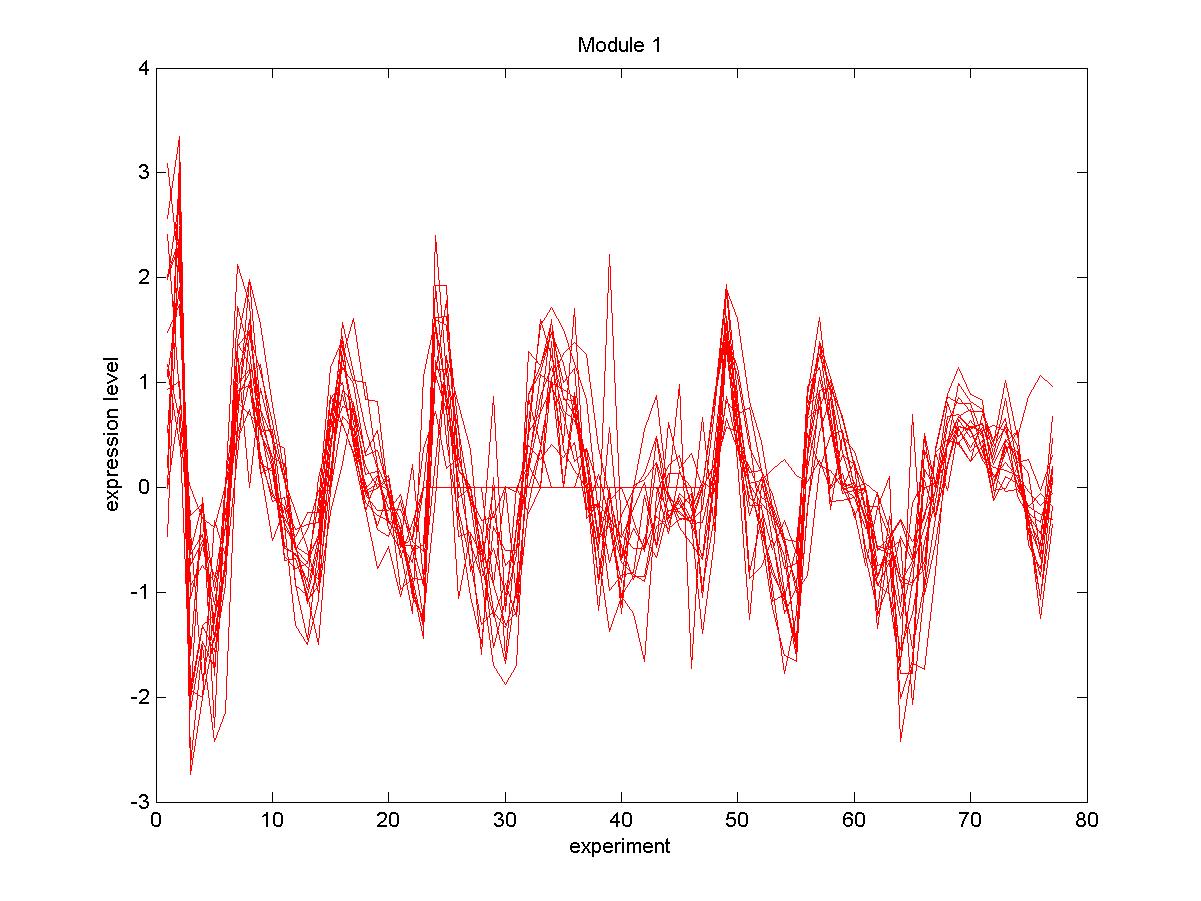
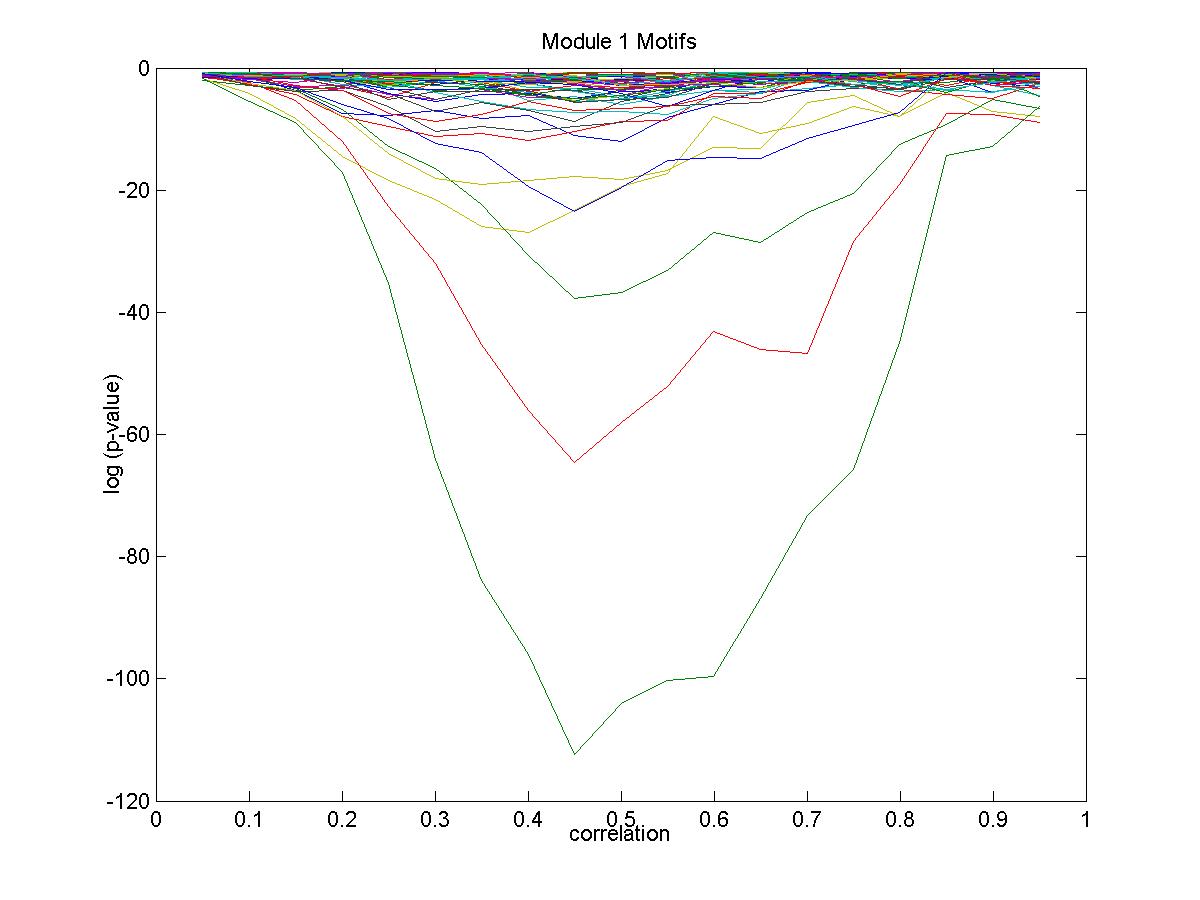
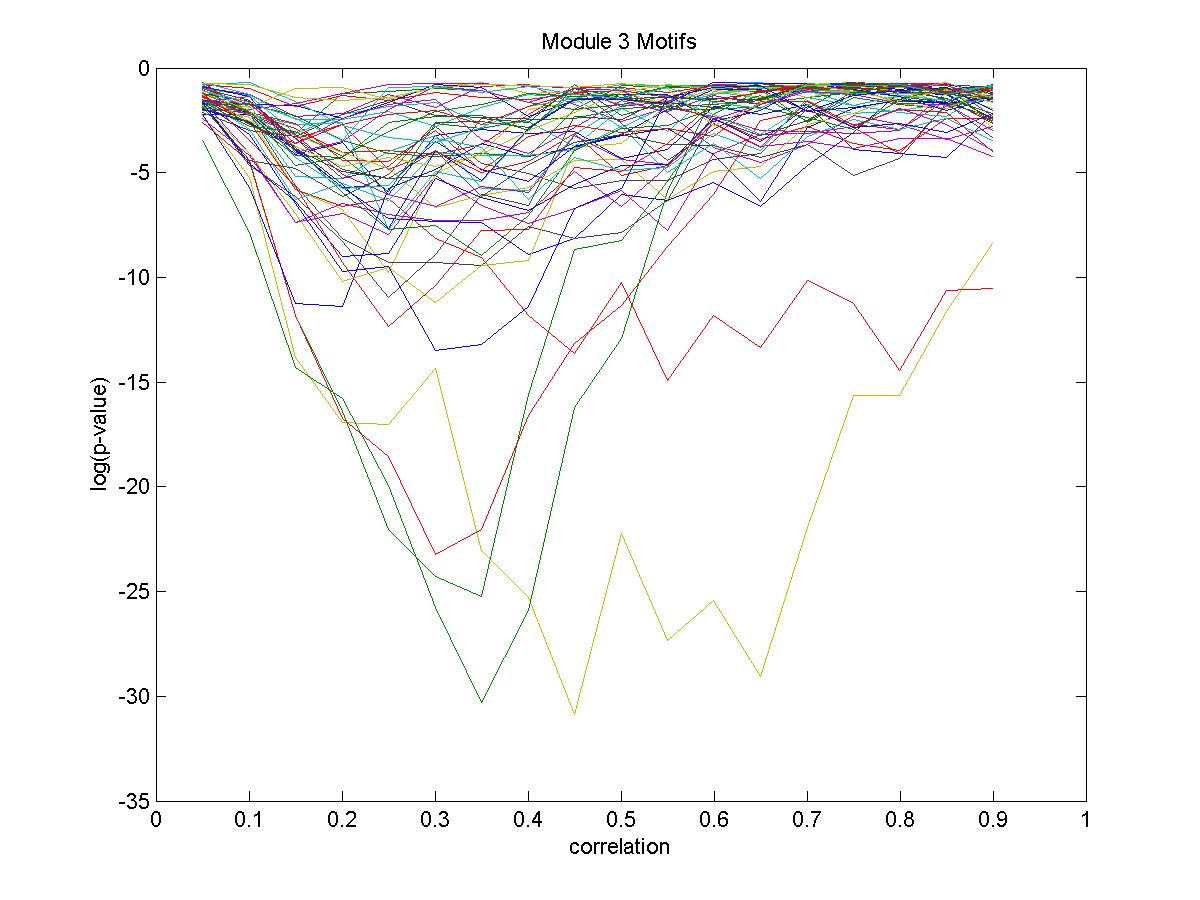
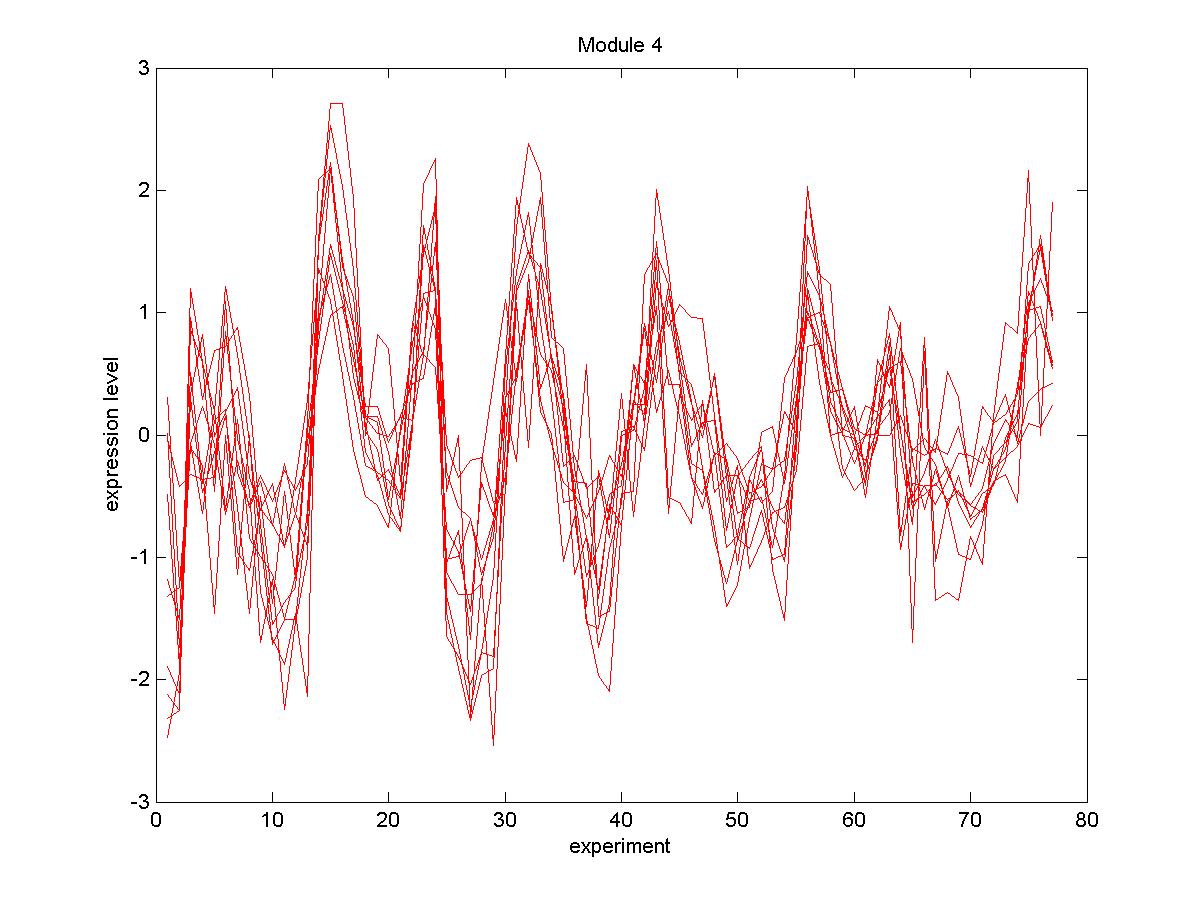
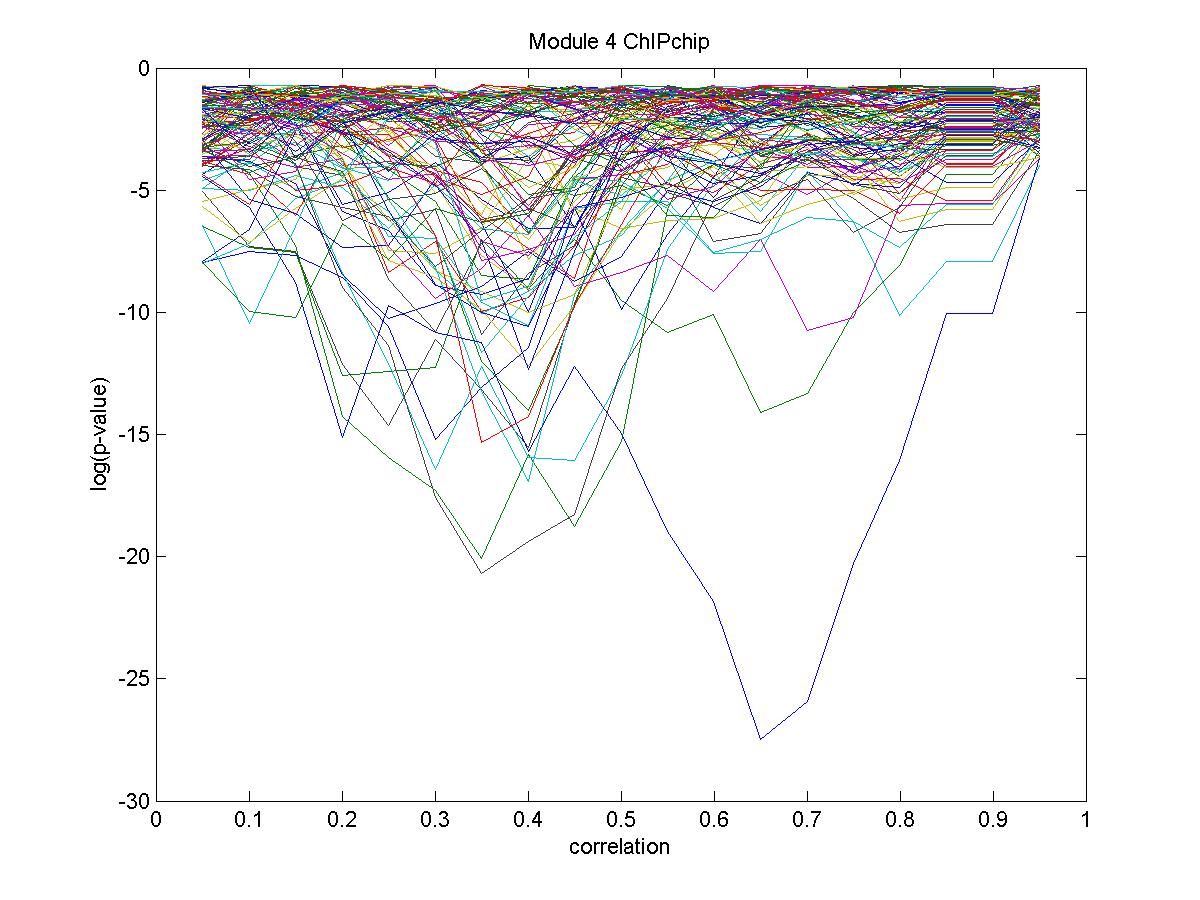
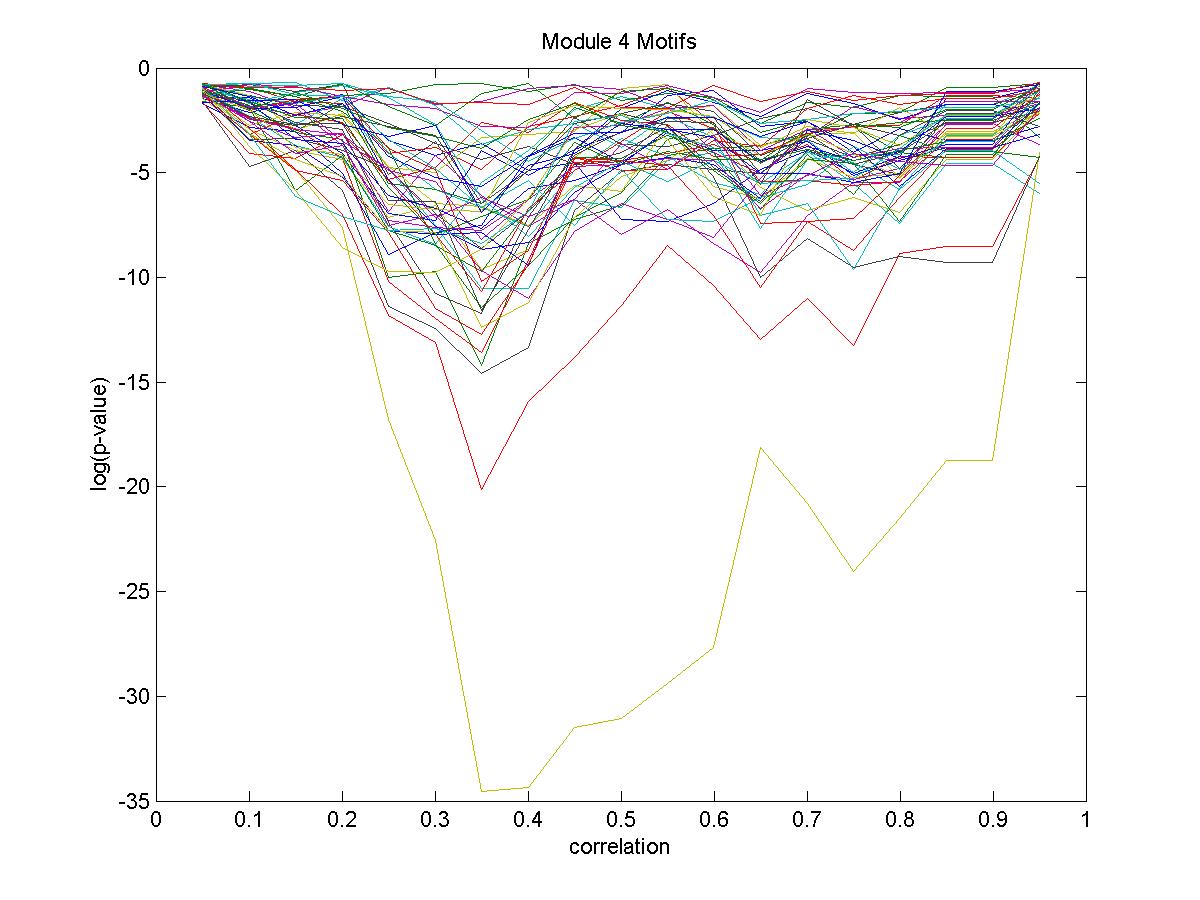
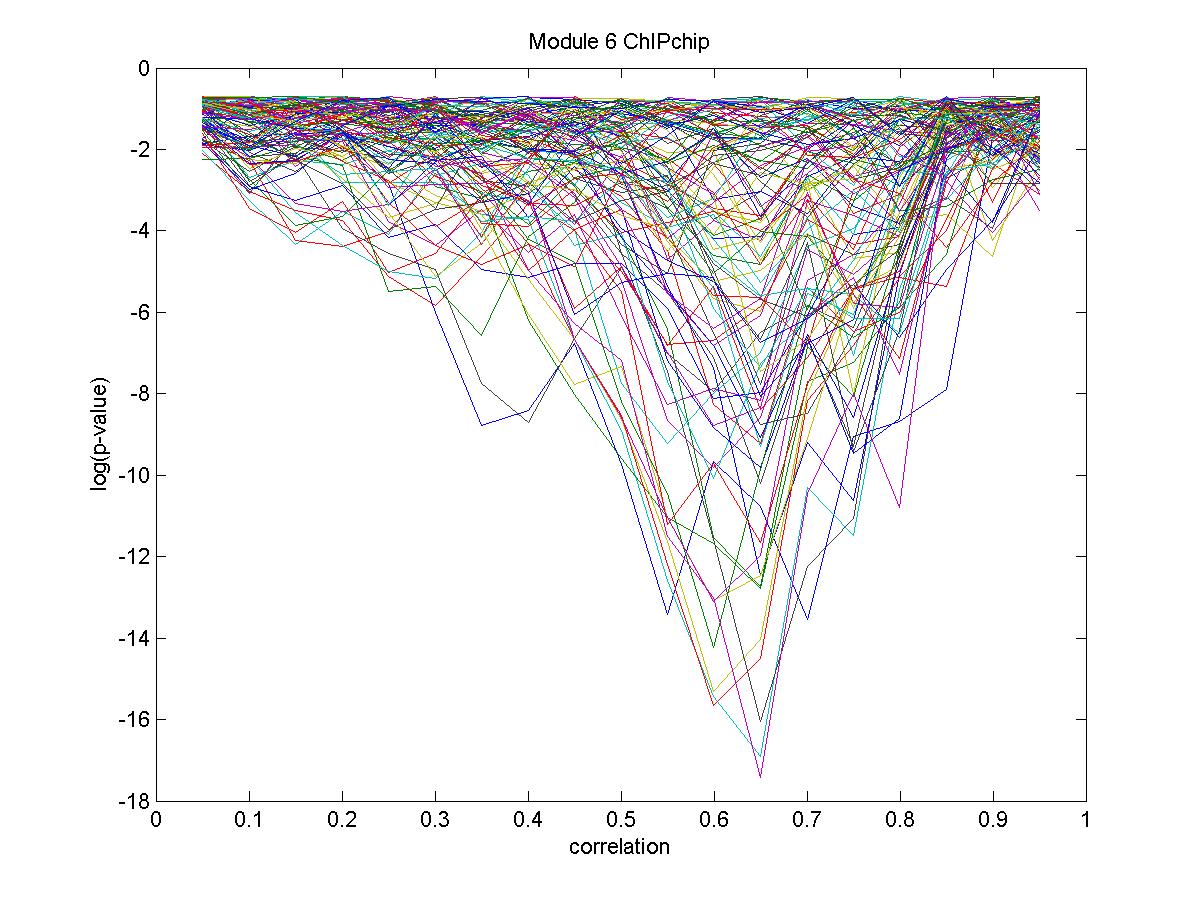
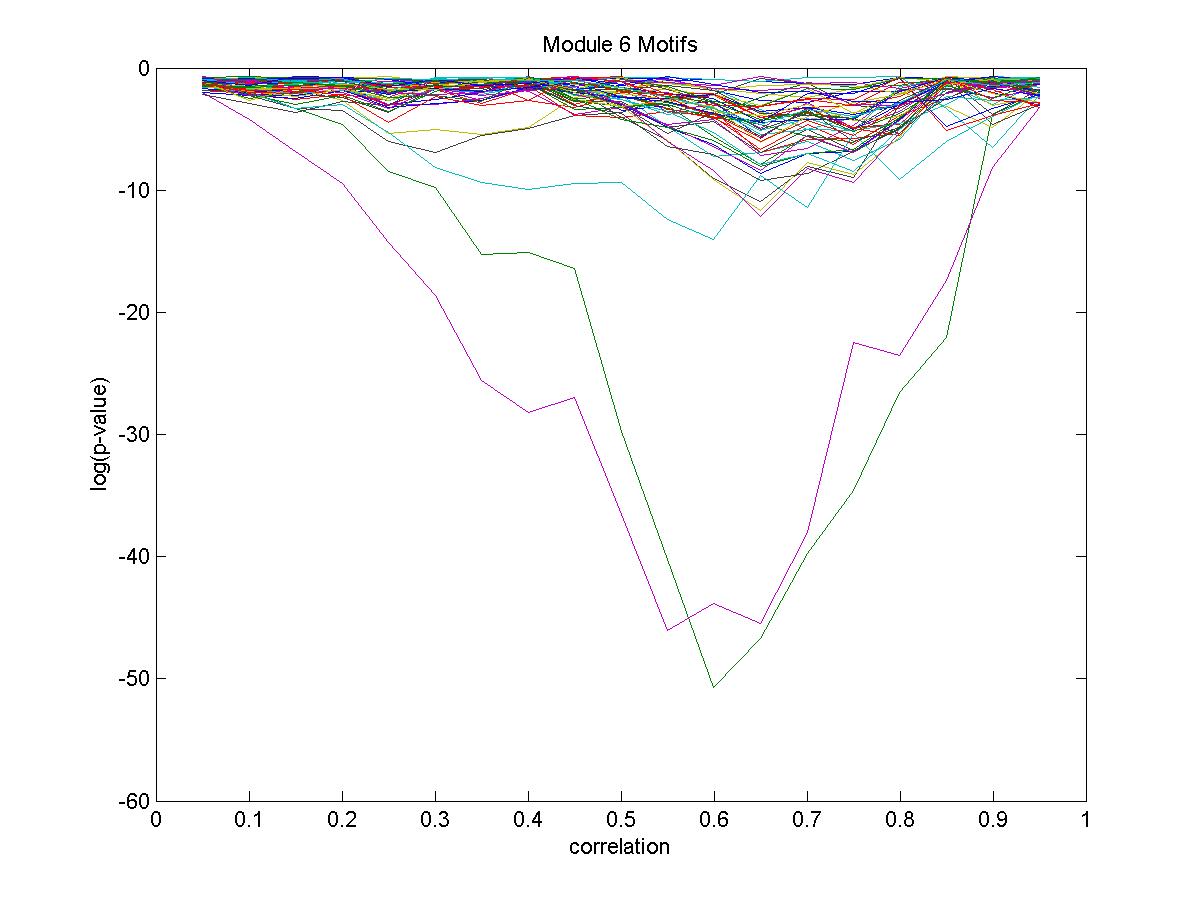
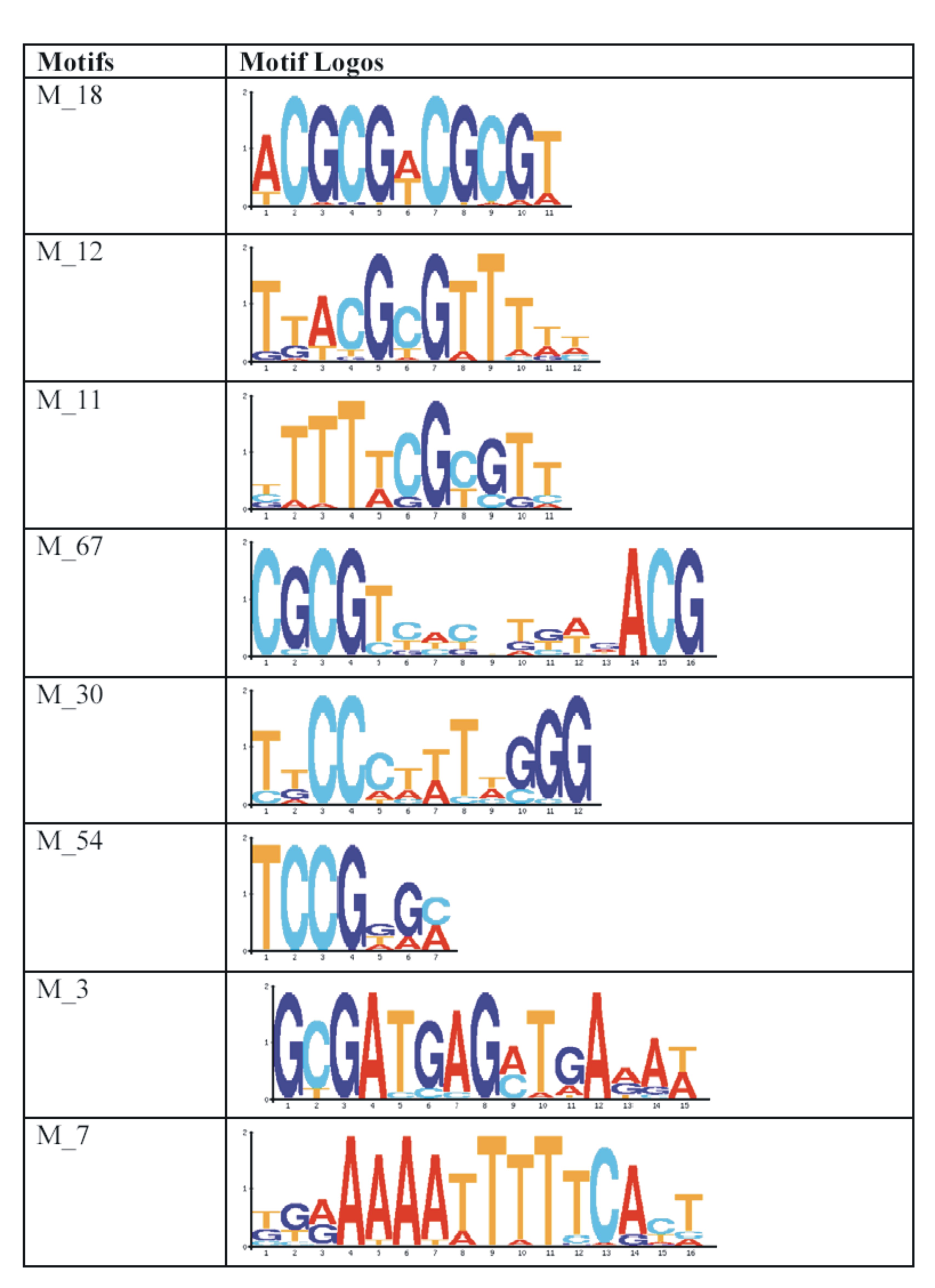
| Gene |
Name |
AccessionNr |
Description |
|---|
| YDL003W |
MCD1 |
NC_001136 |
YDL003W Mcd1p Mitotic Chromosome Determinant; similar to S. pombe RAD21; may function in chromosome morphogenesis from S phase through mitosis |
| YER001W |
MNN1 |
NC_001137 |
YER001W Mnn1p Alpha-1,3-mannosyltransferase |
| YGR109C |
CLB6 |
NC_001139 |
YGR109C Clb6p role in DNA replication during S phase |
| YGR189C |
CRH1 |
NC_001139 |
YGR189C Crh1p congo red hypersensitive |
| YGR221C |
TOS2 |
NC_001139 |
YGR221C Tos2p Hypothetical ORF |
| YHR149C |
YHR149C |
NC_001140 |
Yhr149cp Hypothetical ORF |
| YER070W |
RNR1 |
NC_001137 |
YER070W Rnr1p ribonucleotide reductase |
| YPL256C |
CLN2 |
NC_001148 |
YPL256C Cln2p role in cell cycle START |
| YNL300W |
TOS6 |
NC_001146 |
YNL300W Tos6p Hypothetical ORF |
| YPL163C |
SVS1 |
NC_001148 |
YPL163C Svs1p involved in vanadate resistance |
| YPL267W |
YPL267W |
NC_001148 |
Ypl267wp Hypothetical ORF |
| YPR120C |
CLB5 |
NC_001148 |
YPR120C Clb5p role in DNA replication during S phase; additional functional role in formation of mitotic spindles along with Clb3 and Clb4 |
| YMR199W |
CLN1 |
NC_001145 |
YMR199W Cln1p role in cell cycle START |
| YMR179W |
SPT21 |
NC_001145 |
YMR179W Spt21p involved in trascriptional regulation of Ty1 LTRs |
| YML027W |
YOX1 |
NC_001145 |
YML027W Yox1p Homeodomain protein that binds leu-tRNA gene |
| YKL113C |
RAD27 |
NC_001143 |
YKL113C Rad27p DNA repair protein that belongs to the RAD2(pombe)*FEN1 subfamily |
| Gene |
Name |
AccessionNr |
Description |
|---|
|
|
|
|
| YDL003W |
MCD1 |
NC_001136 |
YDL003W Mcd1p Mitotic Chromosome Determinant; similar to S. pombe RAD21; may function in chromosome morphogenesis from S phase through mitosis |
| YGR143W |
SKN1 |
NC_001139 |
YGR143W Skn1p Involved in (1->6)-beta-glucan biosynthesis |
| YIL123W |
SIM1 |
NC_001141 |
YIL123W Sim1p (putative) invovled in control of DNA replication |
| YHR061C |
GIC1 |
NC_001140 |
YHR061C Gic1p Gtpase-interacting component 1 |
| YDR451C |
YHP1 |
NC_001136 |
YDR451C Yhp1p |
| YBR078W |
ECM33 |
NC_001134 |
YBR078W Ecm33p ExtraCellular Mutant |
| YDR261C |
EXG2 |
NC_001136 |
YDR261C Exg2p Exo-1,3-b-glucanase |
| YJL080C |
SCP160 |
NC_001142 |
YJL080C Scp160p May be required during cell division for faithful partitioning of the ER-nuclear envelope membranes, involved in control of mitotic chromsome transmission |
| YLR300W |
EXG1 |
NC_001144 |
YLR300W Exg1p Exo-1,3-beta-glucanase |
| YLR380W |
CSR1 |
NC_001144 |
YLR380W Csr1p chs5 spa2 rescue; isolated as a multicopy suppressor of the lethality of chs5 spa2 double mutant at 37 degrees. |
| YLR342W |
FKS1 |
NC_001144 |
YLR342W Fks1p Required for viability of calcineurin mutants |
| YLR437C |
YLR437C |
NC_001144 |
Ylr437cp Hypothetical ORF |
| YOR114W |
YOR114W |
NC_001147 |
Yor114wp Hypothetical ORF |
| YPL127C |
HHO1 |
NC_001148 |
YPL127C Hho1p Histone H1 |
| YNL089C |
YNL089C |
NC_001146 |
Ynl089cp Hypothetical ORF |
| YJL187C |
SWE1 |
NC_001142 |
YJL187C Swe1p protein kinase homolog |
| YNR009W |
YNR009W |
NC_001146 |
Ynr009wp Hypothetical ORF |
| YPL163C |
SVS1 |
NC_001148 |
YPL163C Svs1p involved in vanadate resistance |
| YMR199W |
CLN1 |
NC_001145 |
YMR199W Cln1p role in cell cycle START |
| YMR215W |
YMR215W |
NC_001145 |
Ymr215wp Hypothetical ORF |
| YJL186W |
MNN5 |
NC_001142 |
YJL186W Mnn5p mannan synthesis |
| YOR247W |
SRL1 |
NC_001147 |
YOR247W Srl1p Suppressor of rad53 lethality |
| YNL238W |
KEX2 |
NC_001146 |
YNL238W Kex2p prohormone processing; golgi localization marker, dispensable for meiotic recombination but partially required for meiosis I and*or meiosis II |
| YLR056W |
ERG3 |
NC_001144 |
YLR056W Erg3p C-5 sterol desaturase |
| YJL092W |
HPR5 |
NC_001142 |
YJL092W Hpr5p Required for proper timing of committment to meiotic recombination and the transition from Meiosis I to Meiosis II |
| YKL096W |
CWP1 |
NC_001143 |
YKL096W Cwp1p cell wall protein, involved in O and N glycosylation, acceptor of B1-6 glucan. |
| YLR297W |
YLR297W |
NC_001144 |
Ylr297wp Hypothetical ORF |
| YML027W |
YOX1 |
NC_001145 |
YML027W Yox1p Homeodomain protein that binds leu-tRNA gene |
| YJL158C |
CIS3 |
NC_001142 |
YJL158C Cis3p cik1 suppressor |
| Gene |
Name |
AccessionNr |
Description |
|---|
| YDR033W |
MRH1 |
NC_001136 |
YDR033W Mrh1p Membrane protein related to Hsp30p; Localized by immunofluorescence to membranes, mainly the plasma membr. punctuate immunofluorescence pattern observed in buds. The nuclear envelope, but not vacuole or mitochondrial membranes also stained |
| YAR018C |
KIN3 |
NC_001133 |
YAR018C Kin3p protein kinase |
| YDR146C |
SWI5 |
NC_001136 |
YDR146C Swi5p transcriptional activator |
| YIL158W |
YIL158W |
NC_001141 |
Yil158wp Hypothetical ORF |
| YGL116W |
CDC20 |
NC_001139 |
YGL116W Cdc20p Required for onset of anaphase |
| YJL051W |
YJL051W |
NC_001142 |
Yjl051wp Hypothetical ORF |
| YGL021W |
ALK1 |
NC_001139 |
YGL021W Alk1p |
| YGR143W |
SKN1 |
NC_001139 |
YGR143W Skn1p Involved in (1->6)-beta-glucan biosynthesis |
| YHR023W |
MYO1 |
NC_001140 |
YHR023W Myo1p myosin class II |
| YBR038W |
CHS2 |
NC_001134 |
YBR038W Chs2p chitin synthase 2 |
| YCR024C-A |
PMP1 |
NC_001135 |
YCR024C-A Pmp1p May regulate plasma membrane H(+)-ATPase |
| YLR084C |
RAX2 |
NC_001144 |
YLR084C Rax2p Involved in the maintenance of bipolar pattern |
| YMR001C |
CDC5 |
NC_001145 |
YMR001C Cdc5p CDC5 is dispensable for premeiotic DNA synthesis and recombination, but required for tripartite synaptonemal complexes, haploidization, and spores |
| YML119W |
YML119W |
NC_001145 |
Yml119wp Hypothetical ORF |
| YLR190W |
YLR190W |
NC_001144 |
Ylr190wp Hypothetical ORF |
| YPR119W |
CLB2 |
NC_001148 |
YPR119W Clb2p Involved in mitotic induction |
| YPL141C |
YPL141C |
NC_001148 |
Ypl141cp Hypothetical ORF |
| YJR092W |
BUD4 |
NC_001142 |
YJR092W Bud4p co-assembles with Bud3p at bud sites |
| YLR131C |
ACE2 |
NC_001144 |
YLR131C Ace2p involved in transcriptional regulation of CUP1 |
|
|
|
|
| Gene |
Name |
AccessionNr |
Description |
|---|
| YIL009W |
FAA3 |
NC_001141 |
YIL009W Faa3p acyl-CoA synthetase (long-chain fatty acid CoA ligase) (fatty acid activator 3), activates endogenous but not imported fatty acids and provides substrates for N-myristoylation |
| YDL117W |
CYK3 |
NC_001136 |
YDL117W Cyk3p involved in CYtoKinesis |
| YDR055W |
PST1 |
NC_001136 |
YDR055W Pst1p Protoplasts-secreted |
| YJL159W |
HSP150 |
NC_001142 |
YJL159W Hsp150p Heat shock protein, secretory glycoprotein |
| YKL163W |
PIR3 |
NC_001143 |
YKL163W Pir3p Protein containing tandem internal repeats |
| YKL164C |
PIR1 |
NC_001143 |
YKL164C Pir1p Protein containing tandem internal repeats |
| YNL327W |
EGT2 |
NC_001146 |
YNL327W Egt2p cell-cycle regulation protein, may be involved in the correct timing of cell separation after cytokinesis |
| YOR066W |
YOR066W |
NC_001147 |
Yor066wp Hypothetical ORF |
| YNL078W |
JIP1 |
NC_001146 |
YNL078W Jip1p Hypothetical ORF |
| YKL185W |
ASH1 |
NC_001143 |
YKL185W Ash1p Zinc-finger inhibitor of HO transcription which is asymmetrically localized to the daughter cell nucleus |
| Gene |
Name |
AccessionNr |
Description |
|---|
| YDL136W |
RPL35B |
NC_001136 |
YDL136W Rpl35bp Homology to rat L35 |
| YDL130W |
RPP1B |
NC_001136 |
YDL130W Rpp1bp Homology to rat P1, human P1, and E. coli L12eIIB |
| YDL083C |
RPS16B |
NC_001136 |
YDL083C Rps16bp Homology to rat S16 |
| YGR148C |
RPL24B |
NC_001139 |
YGR148C Rpl24bp Homology to rat L24 |
| YDR450W |
RPS18A |
NC_001136 |
YDR450W Rps18ap Homology to rat S18 and E. coli S13 |
| YGL189C |
RPS26A |
NC_001139 |
YGL189C Rps26ap Homology to rat S26 |
| YER056C-A |
RPL34A |
NC_001137 |
YER056C-A Rpl34ap Homology to rat L34 |
| YER131W |
RPS26B |
NC_001137 |
YER131W Rps26bp Homology to rat S26 |
| YGL031C |
RPL24A |
NC_001139 |
YGL031C Rpl24ap Homology to rat L24 |
| YGL103W |
RPL28 |
NC_001139 |
YGL103W Rpl28p Homology to rat, mouse L27a |
| YER102W |
RPS8B |
NC_001137 |
YER102W Rps8bp Homology to mammalian S8 |
| YBL072C |
RPS8A |
NC_001134 |
YBL072C Rps8ap Homology to mammalian S8 |
| YCR031C |
RPS14A |
NC_001135 |
YCR031C Rps14ap Homology to mammalian S14, E. coli S11 |
| YBR118W |
TEF2 |
NC_001134 |
YBR118W Tef2p translational elongation factor EF-1 alpha |
| YBR189W |
RPS9B |
NC_001134 |
YBR189W Rps9bp Homology to rat S9 and E. coli S4 |
| YLR167W |
RPS31 |
NC_001144 |
YLR167W Rps31p Homology to rat S27a |
| YLR061W |
RPL22A |
NC_001144 |
YLR061W Rpl22ap Homology to rat L22 |
| YLR333C |
RPS25B |
NC_001144 |
YLR333C Rps25bp Homology to rat S25; belongs to the S25E family of ribosomal proteins |
| YOL127W |
RPL25 |
NC_001147 |
YOL127W Rpl25p Homology to E. coli L23 and rat L23a |
| YOL040C |
RPS15 |
NC_001147 |
YOL040C Rps15p Homology to rat S15 and E. coli S19 |
| YOR293W |
RPS10A |
NC_001147 |
YOR293W Rps10ap Homology to rat S10 |
| YPL090C |
RPS6A |
NC_001148 |
YPL090C Rps6ap Homology to rat, mouse, and human S6 |
| YPL220W |
RPL1A |
NC_001148 |
YPL220W Rpl1ap Homology to rat L10a, eubacterial L1, and archaebacterial L1; identical to S. cerevisiae L1B (Ssm2p) |
| YOR096W |
RPS7A |
NC_001147 |
YOR096W Rps7ap Homology to human S7 and Xenopus S8 |
| YLR344W |
RPL26A |
NC_001144 |
YLR344W Rpl26ap Homology to rat L26 |
| YLR441C |
RPS1A |
NC_001144 |
YLR441C Rps1ap Homologous to rat S3A |
| Gene |
Name |
AccessionNr |
Description |
|---|
| YCR057C |
PWP2 |
NC_001135 |
YCR057C Pwp2p Eight WD-repeats with homology with G protein beta subunits flanked by nonhomologous N-terminal and C-terminal extensions |
| YCR091W |
KIN82 |
NC_001135 |
YCR091W Kin82p Putative serine*threonine protein kinase most similar to cyclic nucleotide-dependent protein kinase subfamily and the protein kinase C subfamily |
| YDR184C |
ATC1 |
NC_001136 |
YDR184C Atc1p interacts with AIP3, localized to the nucleus |
| YAL025C |
MAK16 |
NC_001133 |
YAL025C Mak16p putative nuclear protein |
| YDR083W |
RRP8 |
NC_001136 |
YDR083W Rrp8p |
| YDR449C |
YDR449C |
NC_001136 |
Ydr449cp Protein required for cell viability |
| YER127W |
LCP5 |
NC_001137 |
YER127W Lcp5p Lethal with conditional pap1 allele |
| YGL099W |
KRE35 |
NC_001139 |
YGL099W Kre35p Killer toxin REsistant |
| YGL171W |
ROK1 |
NC_001139 |
YGL171W Rok1p contains domains found in the DEAD protein family of ATP-dependent RNA helicases; high-copy suppressor of kem1 null mutant |
| YHR196W |
YHR196W |
NC_001140 |
Yhr196wp Protein required for cell viability |
| YIL103W |
YIL103W |
NC_001141 |
Yil103wp Hypothetical ORF |
| YIL127C |
YIL127C |
NC_001141 |
Yil127cp Hypothetical ORF |
| YHR065C |
RRP3 |
NC_001140 |
YHR065C Rrp3p Required for maturation of the 35S primary transcript of pre-rRNA and is required for cleavages leading to mature 18S RNA |
| YGR128C |
YGR128C |
NC_001139 |
Ygr128cp Protein required for cell viability |
| YBR155W |
CNS1 |
NC_001134 |
YBR155W Cns1p cyclophilin seven suppressor |
| YGL120C |
PRP43 |
NC_001139 |
YGL120C Prp43p Pre-mRNA processing factor involved in disassembly of spliceosomes after the release of mature mRNA |
| YGR145W |
YGR145W |
NC_001139 |
Ygr145wp Protein required for cell viability |
| YIL091C |
YIL091C |
NC_001141 |
Yil091cp Protein required for cell viability |
| YER006W |
NUG1 |
NC_001137 |
YER006W Nug1p NUclear GTPase |
| YCR016W |
YCR016W |
NC_001135 |
Ycr016wp Hypothetical ORF |
| YER126C |
NSA2 |
NC_001137 |
YER126C Nsa2p Killer toxin Resistant; Nop seven associated protein 2 |
| YHR085W |
YHR085W |
NC_001140 |
Yhr085wp Protein required for cell viability |
| YAL059W |
ECM1 |
NC_001133 |
YAL059W Ecm1p putative transmembrane domain protein involved in cell wall biogenesis |
| YCL054W |
SPB1 |
NC_001135 |
YCL054W Spb1p Suppressor of PaB1 mutant; involved in 60S ribosomal subunit biogenesis |
| YDR165W |
YDR165W |
NC_001136 |
Ydr165wp Hypothetical ORF |
| YCR072C |
YCR072C |
NC_001135 |
Ycr072cp Protein required for cell viability |
| YKR025W |
RPC37 |
NC_001143 |
YKR025W Rpc37p RNA Polymerase C (III) 37 kDa subunit; interacts with C53 subunit. C37 is an RNA polymerase III specific subunit. |
| YKR081C |
RPF2 |
NC_001143 |
YKR081C Rpf2p Hypothetical ORF |
| YLR196W |
PWP1 |
NC_001144 |
YLR196W Pwp1p Protein with periodic trytophan residues that resembles members of beta-transducin superfamily because of presence of WD-40 repeats |
| YKL099C |
YKL099C |
NC_001143 |
Ykl099cp Protein required for cell viability |
| YLR197W |
SIK1 |
NC_001144 |
YLR197W Sik1p homology to microtubule binding proteins and to X90565_5.cds |
| YMR049C |
ERB1 |
NC_001145 |
YMR049C Erb1p Eukaryotic Ribosome Biogenesis |
| YKL172W |
EBP2 |
NC_001143 |
YKL172W Ebp2p EBNA1-binding protein homolog |
| YOR001W |
RRP6 |
NC_001147 |
YOR001W Rrp6p Ribosomal RNA Processing |
| YML093W |
YML093W |
NC_001145 |
Yml093wp Protein required for cell viability |
| YNL248C |
RPA49 |
NC_001146 |
YNL248C Rpa49p 49-kDa alpha subunit of RNA polymerase A |
| YOL144W |
NOP8 |
NC_001147 |
YOL144W Nop8p Nucleolar protein required for 60S ribosome biogenesis |
| YNR046W |
YNR046W |
NC_001146 |
Ynr046wp Protein required for cell viability |
| YOL041C |
NOP12 |
NC_001147 |
YOL041C Nop12p Nucleolar Protein; isolated as a mutant exhibiting synthetic lethality with a nop2 ts allele. |
| YOR091W |
YOR091W |
NC_001147 |
Yor091wp Hypothetical ORF |
| YPL217C |
BMS1 |
NC_001148 |
YPL217C Bms1p BMH1 sensitive |
| YMR269W |
YMR269W |
NC_001145 |
Ymr269wp protein possibly involved in protein synthesis |
| YOR277C |
YOR277C |
NC_001147 |
Yor277cp Hypothetical ORF |
| YOR340C |
RPA43 |
NC_001147 |
YOR340C Rpa43p DNA-dependent RNA polymerase I subunit A43 |
| YNL132W |
KRE33 |
NC_001146 |
YNL132W Kre33p Killer toxin REsistant |
| YOR206W |
NOC2 |
NC_001147 |
YOR206W Noc2p NucleOlar Complex 2; involved in nuclear export of pre-ribosomes |
| YPL211W |
NIP7 |
NC_001148 |
YPL211W Nip7p Nip7p is required for 60S ribosome subunit biogenesis |
| YNL061W |
NOP2 |
NC_001146 |
YNL061W Nop2p May participate in nucleolar function during the transition from stationary phase to rapid growth |
| YNL110C |
NOP15 |
NC_001146 |
YNL110C Nop15p Nucleolar protein 15 |
| YJR002W |
MPP10 |
NC_001142 |
YJR002W Mpp10p Protein component of the U3 small nucleolar ribonucleoprotein (snoRNP) |
| YMR128W |
ECM16 |
NC_001145 |
YMR128W Ecm16p ExtraCellular Mutant |
| YLR129W |
DIP2 |
NC_001144 |
YLR129W Dip2p DOM34 Interacting Protein |
| YMR014W |
BUD22 |
NC_001145 |
YMR014W Bud22p |
| YJL109C |
YJL109C |
NC_001142 |
Yjl109cp Protein required for cell viability |
| YKL144C |
RPC25 |
NC_001143 |
YKL144C Rpc25p Subunit of RNA polymerase III |
| YLR074C |
BUD20 |
NC_001144 |
YLR074C Bud20p |
| YLR449W |
FPR4 |
NC_001144 |
YLR449W Fpr4p Homolog of homolog of the nucleolar FKBP, Fpr3 |
| YKL082C |
YKL082C |
NC_001143 |
Ykl082cp Protein required for cell viability |
| YKR024C |
DBP7 |
NC_001143 |
YKR024C Dbp7p Dead-box protein |
| YLR051C |
YLR051C |
NC_001144 |
Ylr051cp Protein required for cell viability |