Tessellation and Interpolation of Scattered Data
This demo describes convex hulls, Delaunay tessellations, and Voronoi diagrams in 3 dimensions. It also shows how to interpolate three-dimensional scattered data.
Many applications in science, engineering, statistics, and mathematics use structures like convex hulls, Delaunay tessellations and Voronoi diagrams for analyzing data. MATLAB® enables you to geometrically analyze data sets in any dimension.
Here, in 3 dimensions, we show a set of 50 points with its convex hull.
% Create the data. n = 50; X = randn(n,3); % Plot the points. plot3(X(:,1),X(:,2),X(:,3),'ko','markerfacecolor','k'); % Compute the convex hull. C = convhulln(X); % Plot the convex hull. hold on for i = 1:size(C,1) j = C(i,[1 2 3 1]); patch(X(j,1),X(j,2),X(j,3),rand,'FaceAlpha',0.6); end % Modify the view. view(3), axis equal off tight vis3d; camzoom(1.2) colormap(spring) rotate3d on

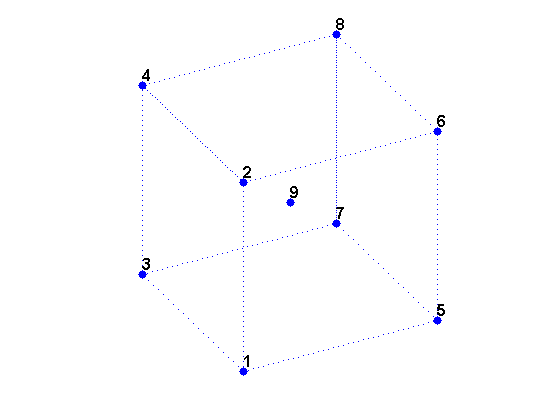
We can create a data set X of the 8 vertices of a cube plus its center.
X is a 9-by-3 matrix where each row is the 3-D coordinates of one point.
% Create X. X = zeros(8,3); X([5:8,11,12,15,16,18,20,22,24]) = 1; % Corners. X(9,:) = [0.5 0.5 0.5]; % Center. % Visualize X. cla reset; hold on d = [1 2 4 3 1 5 6 8 7 5 6 2 4 8 7 3]; plot3(X(d,1),X(d,2),X(d,3),'b:'); plot3(X(:,1),X(:,2),X(:,3),'b.','markersize',20); t = text(X(:,1),X(:,2),X(:,3), num2str((1:9)')); set(t,'VerticalAlignment','bottom','FontWeight','bold', 'FontSize',12); view(3); axis equal tight off vis3d; camorbit(10,0); rotate3d on
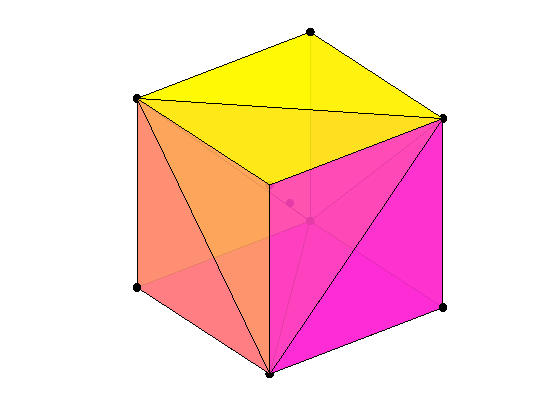
The convex hull of a data set is the smallest convex region that contains the data set. The convex hull of the cube data set X can be computed by CONVHULLN.
For this data set X, the convex hull has 12 facets, each corresponding to a row in K and plotted above with X. The cube is transparent so that you can see all the facets and data points.
% Compute the convex hull. tri = convhulln(X); % Plot the data cla reset; plot3(X(:,1),X(:,2),X(:,3),'ko','markerfacecolor','k'); % Plot the convex hull. for i = 1:size(tri,1) c = tri(i,[1 2 3 1]); patch(X(c,1),X(c,2),X(c,3),i,'FaceAlpha', 0.9); end % Modify the view. view(3); axis equal tight off vis3d rotate3d on
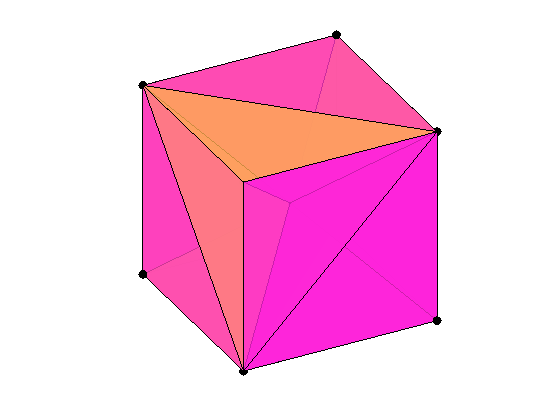
A Delaunay tessellation in 3 dimensions is a set of tetrahedrons such that no data points are contained in any tetrahedron's circumsphere. The Delaunay tessellation of the data set X can be computed by DELAUNAYN.
The 12 rows of T represent the 12 tetrahedrons that partition the data set X.
% Compute the delaunay tessellation. tri = delaunayn(X); % Plot the data. plot3(X(:,1),X(:,2),X(:,3),'ko','markerfacecolor','k'); % Plot the tessellation. for i = 1:size(tri,1) y = tri(i,[1 1 1 2; 2 2 3 3; 3 4 4 4]); x1 = reshape(X(y,1),3,4); x2 = reshape(X(y,2),3,4); x3 = reshape(X(y,3),3,4); patch(x1,x2,x3,(1:4)*i,'FaceAlpha',0.8); end % Modify the view. view(3); axis equal tight off vis3d; camorbit(10,0) rotate3d on
A Voronoi diagram partitions the data space into polyhedral regions, with one region for each data point. Anywhere within a region is closer to its data point than any other in the set. The Voronoi diagram of the cube data set X can be computed by VORONOIN.
V is the set of Voronoi vertices. C represents the set of Voronoi regions. For our data set X, C has 9 Voronoi regions. Here we show one Voronoi region, the region for the center point of the cube.
% Compute Voronoi diagram. [c,v] = voronoin(X); % Plot the data. plot3(X(d,1),X(d,2),X(d,3),'b:.',X(9,1),X(9,2),X(9,3),'k.','markersize',20); % Plot the Voronoi diagram. nx = c(v{9},:); tri = convhulln(nx); for i = 1:size(tri,1) patch(nx(tri(i,:),1),nx(tri(i,:),2),nx(tri(i,:),3),rand,'FaceAlpha',0.8); end % Modify the view. view(3); axis equal tight off vis3d; camzoom(1.5); camorbit(20,0) rotate3d on

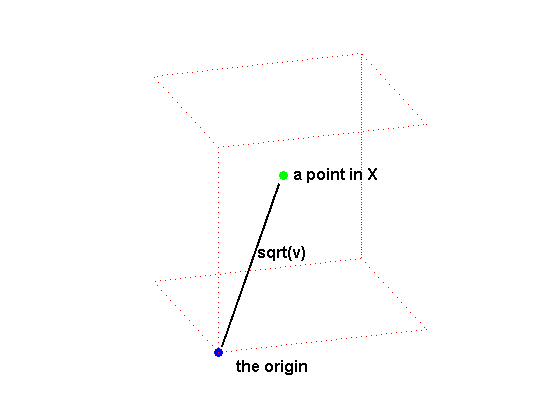
GRIDDATAN interpolates multidimensional scattered data. It uses DELAUNAYN to tessellate the data, and then interpolates based on the tessellation. We start with a data set of 500 random points in 3 dimensions and compute the values of a function, the squared distance from the origin, at each of these points.
% Create the data. n = 500; X = 2*rand(n,3)-1; v = sum(X.^2,2); % Draw a picture to show how X is defined. cla reset; hold on plot3([0.02, 0.47],[0.02,0.57],[0.02,0.57],'k-','linewidth',2); plot3(0,0,0,'bo','markerfacecolor','b'); cube = zeros(8,3); cube([5:8,11,12,15,16,18,20,22,24]) = 1; % Corners cube(9,:) = [0.5 0.5 0.5]; % Center. plot3(cube(d,1),cube(d,2),cube(d,3),'r:'); plot3(0.5,0.6,0.6,'go','markerfacecolor','g'); text(0.02,-0.2,0,'the origin','fontsize',12,'fontweight','bold'); text(0.55,0.6,0.6,'a point in X','fontsize',12,'fontweight','bold'); text(0.28,0.3,0.35,'sqrt(v)','fontsize',12,'fontweight','bold'); view(3); axis equal tight off vis3d; camorbit(20,-10); rotate3d on
With GRIDDATAN we can interpolate X and the values v over a grid X0 to get the function values v0 over this grid.
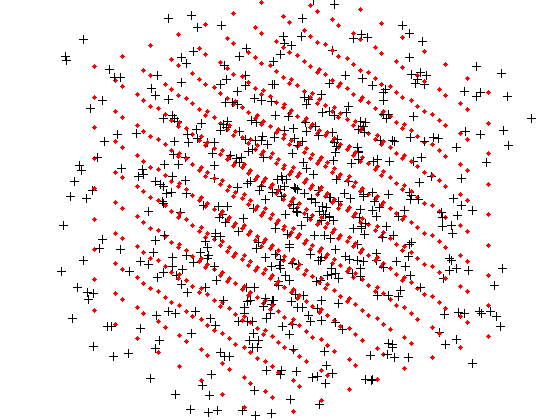
The black points are X and the red points are the grid X0.
% Grid the data. d = -0.8:0.2:0.8; [x0,y0,z0] = meshgrid(d,d,d); X0 = [x0(:) y0(:) z0(:)]; v0 = reshape(griddatan(X,v,X0),size(x0)); % Plot results. cla reset; hold on; plot3(X(:,1),X(:,2),X(:,3),'k+','markerfacecolor','k'); plot3(X0(:,1),X0(:,2),X0(:,3),'r.','markerfacecolor','r'); view(3); axis equal tight off vis3d; camzoom(1.6); rotate3d on
To visualize the surface at all points where the function takes on a constant value, we can use ISOSURFACE and ISONORMALS. Since the function is the squared distance from the origin, the surface at a constant value is a sphere.
c = 0.6; % constant value cla reset; hold on; h = patch(isosurface(x0,y0,z0,v0,c),'FaceColor','red','EdgeColor','none'); isonormals(x0,y0,z0,v0,h); view(3); axis equal tight off vis3d; camzoom(1.6); camlight; lighting phong rotate3d on
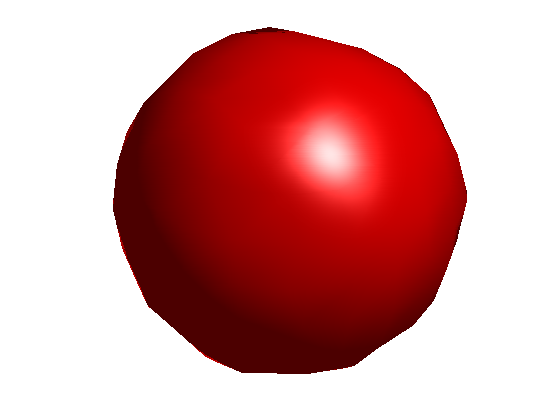
With more data points in X and a denser grid X0, the sphere is smoother but takes longer to compute.
Here is a precomputed sphere generated using 5000 data points in X and a distance between gridpoints of 0.05.
% Load saved results. load qhulldemo cla reset; hold on d = -0.8:0.05:0.8; [x0,y0,z0] = meshgrid(d,d,d); h = patch(isosurface(x0,y0,z0,v0,0.6)); isonormals(x0,y0,z0,v0,h); set(h,'FaceColor','red','EdgeColor','none'); view(3); axis equal tight off vis3d; camzoom(1.6) camlight; lighting phong rotate3d on
